3ZLN
 
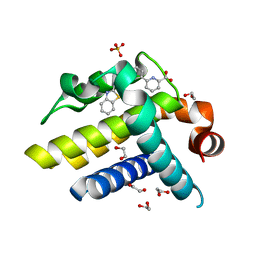 | | Crystal structure of BCL-XL in complex with inhibitor (Compound 3) | | Descriptor: | 1,2-ETHANEDIOL, 6-[(8E)-8-(1,3-benzothiazol-2-ylhydrazinylidene)-6,7-dihydro-5H-naphthalen-2-yl]pyridine-2-carboxylic acid, BCL-2-LIKE PROTEIN 1, ... | | Authors: | Czabotar, P.E, Lessene, G.L, Smith, B.J, Colman, P.M. | | Deposit date: | 2013-02-04 | | Release date: | 2013-04-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.288 Å) | | Cite: | Structure-Guided Design of a Selective Bcl-Xl Inhibitor
Nat.Chem.Biol., 9, 2013
|
|
4A1U
 
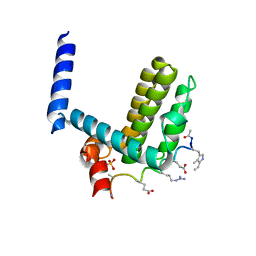 | | Crystal structure of alpha-beta-foldamer 2c in complex with Bcl-xL | | Descriptor: | ALPHA-BETA-FOLDAMER 2C, BCL-2-LIKE PROTEIN 1, CHLORIDE ION, ... | | Authors: | Boersma, M.D, Haase, H.S, Kaufman, K.J, Horne, W.S, Lee, E.F, Clarke, O.B, Smith, B.J, Colman, P.M, Gellman, S.H, Fairlie, W.D. | | Deposit date: | 2011-09-20 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Evaluation of Diverse Alpha/Beta-Backbone Patterns for Functional Alpha-Helix Mimicry: Analogues of the Bim Bh3 Domain.
J.Am.Chem.Soc., 134, 2012
|
|
4A1W
 
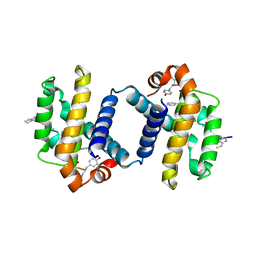 | | Crystal structure of alpha-beta foldamer 4c in complex with Bcl-xL | | Descriptor: | ALPHA-BETA-FOLDAMER 2C, BCL-2-LIKE PROTEIN 1 | | Authors: | Boersma, M.D, Haase, H.S, Kaufman, K.J, Horne, W.S, Lee, E.F, Clarke, O.B, Smith, B.J, Colman, P.M, Gellman, S.H, Fairlie, W.D. | | Deposit date: | 2011-09-20 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Evaluation of Diverse Alpha/Beta-Backbone Patterns for Functional Alpha-Helix Mimicry: Analogues of the Bim Bh3 Domain.
J.Am.Chem.Soc., 134, 2012
|
|
2WLK
 
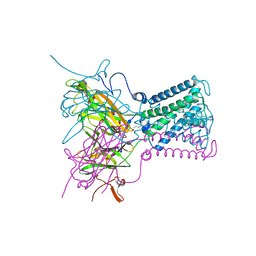 | | STRUCTURE OF THE ATP-SENSITIVE INWARD RECTIFIER POTASSIUM CHANNEL FROM MAGNETOSPIRILLUM MAGNETOTACTICUM | | Descriptor: | ATP-SENSITIVE INWARD RECTIFIER POTASSIUM CHANNEL 10, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Clarke, O.B, Caputo, A.T, Smith, B.J, Gulbis, J.M. | | Deposit date: | 2009-06-24 | | Release date: | 2010-06-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Domain Reorientation and Rotation of an Intracellular Assembly Regulate Conduction in Kir Potassium Channels.
Cell(Cambridge,Mass.), 141, 2010
|
|
2WLJ
 
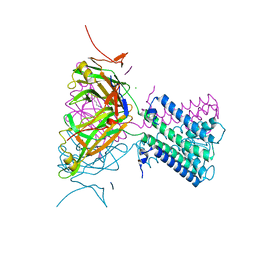 | | Potassium channel from Magnetospirillum magnetotacticum | | Descriptor: | CALCIUM ION, CHLORIDE ION, POTASSIUM CHANNEL, ... | | Authors: | Clarke, O.B, Caputo, A.T, Smith, B.J, Gulbis, J.M. | | Deposit date: | 2009-06-24 | | Release date: | 2010-06-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Domain Reorientation and Rotation of an Intracellular Assembly Regulate Conduction in Kir Potassium Channels.
Cell(Cambridge,Mass.), 141, 2010
|
|
4ZL4
 
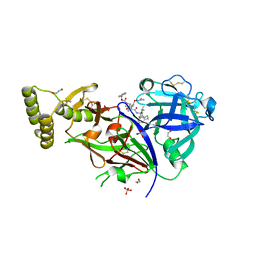 | | Plasmepsin V from Plasmodium vivax bound to a transition state mimetic (WEHI-842) | | Descriptor: | (2R)-1-[(2R)-2-(2-methoxyethoxy)propoxy]propan-2-amine, 1,2-ETHANEDIOL, Aspartic protease PM5, ... | | Authors: | Czabotar, P.E, Hodder, A.N, Smith, B.J, Sleebs, B.E, Gazdic, M, Boddey, J.A, Cowman, A.F. | | Deposit date: | 2015-05-01 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural basis for plasmepsin V inhibition that blocks export of malaria proteins to human erythrocytes.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4ZXB
 
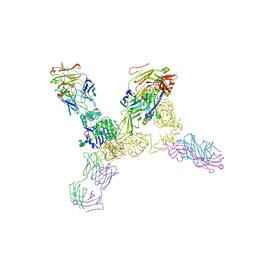 | | Structure of the human insulin receptor ectodomain, IRDeltabeta construct, in complex with four Fab molecules | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Croll, T, Smith, B.J, Margetts, M.B, Whittaker, J, Weiss, M.A, Ward, C.W, Lawrence, M.C. | | Deposit date: | 2015-05-20 | | Release date: | 2016-02-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Higher-Resolution Structure of the Human Insulin Receptor Ectodomain: Multi-Modal Inclusion of the Insert Domain.
Structure, 24, 2016
|
|
1FDZ
 
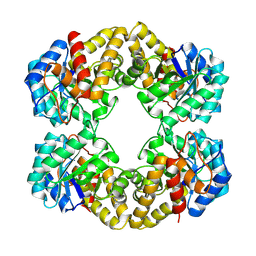 | | N-ACETYLNEURAMINATE LYASE IN COMPLEX WITH PYRUVATE VIA BOROHYDRIDE REDUCTION | | Descriptor: | N-ACETYLNEURAMINATE LYASE, PYRUVIC ACID | | Authors: | Lawrence, M.C, Barbosa, J.A.R.G, Smith, B.J, Hall, N.E, Pilling, P.A, Ooi, H.C, Marcuccio, S.M. | | Deposit date: | 1996-07-08 | | Release date: | 1997-10-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and mechanism of a sub-family of enzymes related to N-acetylneuraminate lyase.
J.Mol.Biol., 266, 1997
|
|
1FDY
 
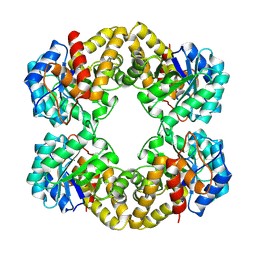 | | N-ACETYLNEURAMINATE LYASE IN COMPLEX WITH HYDROXYPYRUVATE | | Descriptor: | 3-HYDROXYPYRUVIC ACID, N-ACETYLNEURAMINATE LYASE | | Authors: | Lawrence, M.C, Barbosa, J.A.R.G, Smith, B.J, Hall, N.E, Pilling, P.A, Ooi, H.C, Marcuccio, S.M. | | Deposit date: | 1996-07-08 | | Release date: | 1997-10-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure and mechanism of a sub-family of enzymes related to N-acetylneuraminate lyase.
J.Mol.Biol., 266, 1997
|
|
1G5G
 
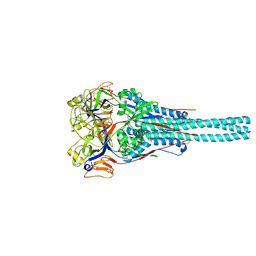 | | FRAGMENT OF FUSION PROTEIN FROM NEWCASTLE DISEASE VIRUS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0, ... | | Authors: | Lawrence, M.C, Smith, B.J. | | Deposit date: | 2000-11-01 | | Release date: | 2002-02-27 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The structure of the fusion glycoprotein of Newcastle disease virus suggests a novel paradigm for the molecular mechanism of membrane fusion.
Structure, 9, 2001
|
|
6GF3
 
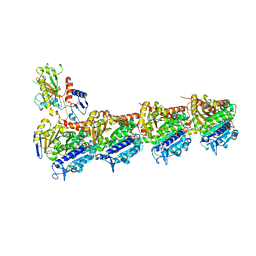 | | Tubulin-Jerantinine B acetate complex | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Smedley, C.J, Stanley, P.A, Qazzaz, M.E, Prota, A.E, Olieric, N, Collins, H, Eastman, H, Barrow, A.S, Lim, K.-H, Kam, T.-S, Smith, B.J, Duivenvoorden, H.M, Parker, B.S, Bradshaw, T.D, Steinmetz, M.O, Moses, J.E. | | Deposit date: | 2018-04-29 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Sustainable Syntheses of (-)-Jerantinines A & E and Structural Characterisation of the Jerantinine-Tubulin Complex at the Colchicine Binding Site.
Sci Rep, 8, 2018
|
|
5FMJ
 
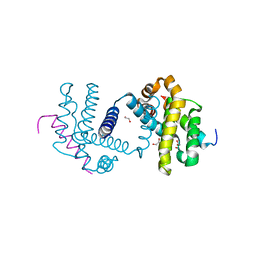 | | Bcl-xL with mouse Bak BH3 Q75L complex | | Descriptor: | 1,2-ETHANEDIOL, BAK1 PROTEIN, BCL-2-LIKE PROTEIN 1 | | Authors: | Fairlie, W.D, Lee, E.F, Smith, B.J. | | Deposit date: | 2015-11-06 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Physiological Restraint of Bak by Bcl-Xl is Essential for Cell Survival.
Genes Dev., 30, 2016
|
|
5FMK
 
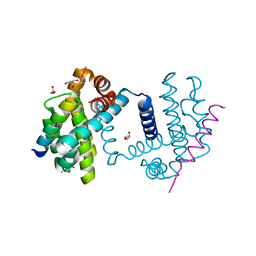 | | Bcl-xL with Bak BH3 complex | | Descriptor: | BCL-2 HOMOLOGOUS ANTAGONIST/KILLER, BCL-XL, GLYCEROL | | Authors: | Fairlie, W.D, Lee, E.F, Smith, B.J, Czabotar, P.E, Colman, P.M. | | Deposit date: | 2015-11-06 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.731 Å) | | Cite: | Physiological Restraint of Bak by Bcl-Xl is Essential for Cell Survival.
Genes Dev., 30, 2016
|
|
5FMI
 
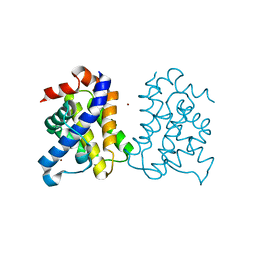 | | Human Bak Q77L | | Descriptor: | BCL-2 HOMOLOGOUS ANTAGONIST/KILLER, ZINC ION | | Authors: | Fairlie, W.D, Lee, E.F, Smith, B.J. | | Deposit date: | 2015-11-06 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.491 Å) | | Cite: | Physiological Restraint of Bak by Bcl-Xl is Essential for Cell Survival.
Genes Dev., 30, 2016
|
|
6X44
 
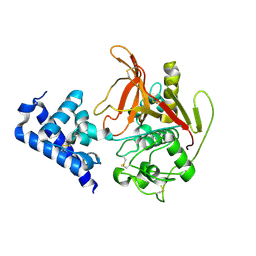 | | High Resolution Crystal Structure Analysis of SERA5 proenzyme from plasmodium falciparum | | Descriptor: | DI(HYDROXYETHYL)ETHER, Serine repeat antigen 5 | | Authors: | Clarke, O.B, Smith, N.A, Lee, M, Smith, B.J. | | Deposit date: | 2020-05-22 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.19733953 Å) | | Cite: | Structure of the Plasmodium falciparum PfSERA5 pseudo-zymogen.
Protein Sci., 29, 2020
|
|
6X42
 
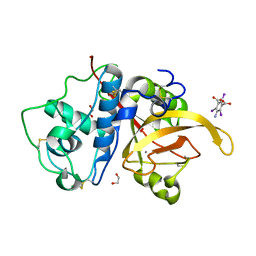 | | High Resolution Crystal Structure Analysis of SERA5E from plasmodium falciparum | | Descriptor: | 1,2-ETHANEDIOL, 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, CALCIUM ION, ... | | Authors: | Clarke, O.B, Smith, N.A, Lee, M, Smith, B.J. | | Deposit date: | 2020-05-21 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of the Plasmodium falciparum PfSERA5 pseudo-zymogen.
Protein Sci., 29, 2020
|
|
1IEX
 
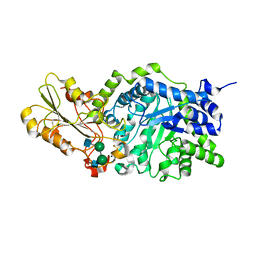 | | Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme Exo1 in complex with 4I,4III,4V-S-trithiocellohexaose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Hrmova, M, DeGori, R, Fincher, G.B, Smith, B.J, Driguez, H, Varghese, J.N. | | Deposit date: | 2001-04-11 | | Release date: | 2001-11-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Catalytic mechanisms and reaction intermediates along the hydrolytic pathway of a plant beta-D-glucan glucohydrolase.
Structure, 9, 2001
|
|
1J8V
 
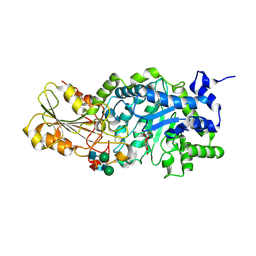 | | Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme Exo1 in complex with 4'-nitrophenyl 3I-thiolaminaritrioside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4'-NITROPHENYL-S-(BETA-D-GLUCOPYRANOSYL)-(1-3)-(3-THIO-BETA-D-GLUCOPYRANOSYL)-(1-3)-BETA-D-GLUCOPYRANOSIDE, ... | | Authors: | Hrmova, M, De Gori, R, Smith, B.J, Fairweather, J.K, Driguez, H, Varghese, J.N, Fincher, G.B. | | Deposit date: | 2001-05-22 | | Release date: | 2002-06-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for broad substrate specificity in higher plant beta-D-glucan glucohydrolases.
Plant Cell, 14, 2002
|
|
1IEW
 
 | | Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme Exo1 in complex with 2-deoxy-2-fluoro-alpha-D-glucoside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-deoxy-2-fluoro-alpha-D-glucopyranose, ... | | Authors: | Hrmova, M, DeGori, R, Fincher, G.B, Smith, B.J, Varghese, J.N. | | Deposit date: | 2001-04-11 | | Release date: | 2001-11-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Catalytic mechanisms and reaction intermediates along the hydrolytic pathway of a plant beta-D-glucan glucohydrolase.
Structure, 9, 2001
|
|
1LQ2
 
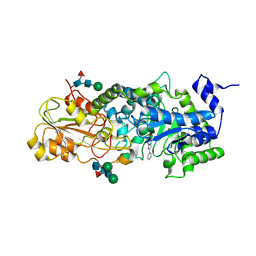 | | Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme Exo1 in complex with gluco-phenylimidazole | | Descriptor: | (5R,6R,7S,8S)-5-(HYDROXYMETHYL)-2-PHENYL-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hrmova, M, De Gori, R, Smith, B.J, Vasella, A, Varghese, J.N, Fincher, G.B. | | Deposit date: | 2002-05-09 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Three-dimensional Structure of the Barley {beta}-D-Glucan Glucohydrolase in Complex with a Transition State Mimic.
J.Biol.Chem., 279, 2004
|
|
2JM6
 
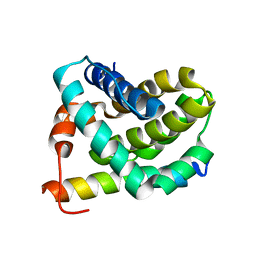 | | Solution structure of MCL-1 complexed with NOXAB | | Descriptor: | Myeloid cell leukemia-1 protein Mcl-1 homolog, Noxa | | Authors: | Czabotar, P.E, Lee, E.F, van Delft, M.F, Day, C.L, Smith, B.J, Huang, D.C.S, Fairlie, W.D, Hinds, M.G, Colman, P.M. | | Deposit date: | 2006-10-17 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the degradation of Mcl-1 induced by BH3 domains
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2X6A
 
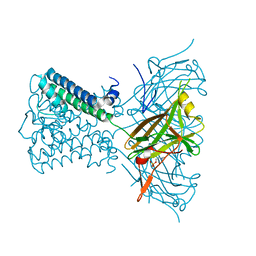 | | Potassium Channel from Magnetospirillum Magnetotacticum | | Descriptor: | ATP-SENSITIVE INWARD RECTIFIER POTASSIUM CHANNEL 10, PHOSPHOCHOLINE, POTASSIUM ION | | Authors: | Clarke, O.B, Caputo, A.T, Hill, A.P, Vandenberg, J.I, Smith, B.J, Gulbis, J.M. | | Deposit date: | 2010-02-15 | | Release date: | 2010-06-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Domain Reorientation and Rotation of an Intracellular Assembly Regulate Conduction in Kir Potassium Channels.
Cell(Cambridge,Mass.), 141, 2010
|
|
2WLN
 
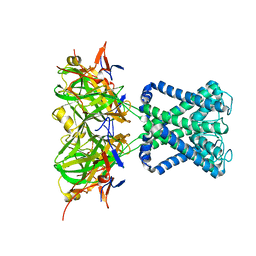 | | POTASSIUM CHANNEL FROM MAGNETOSPIRILLUM MAGNETOTACTICUM | | Descriptor: | POTASSIUM CHANNEL, POTASSIUM ION | | Authors: | Clarke, O.B, Caputo, A.T, Smith, B.J, Gulbis, J.M. | | Deposit date: | 2009-06-24 | | Release date: | 2010-06-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.44 Å) | | Cite: | Domain Reorientation and Rotation of an Intracellular Assembly Regulate Conduction in Kir Potassium Channels.
Cell(Cambridge,Mass.), 141, 2010
|
|
2WLO
 
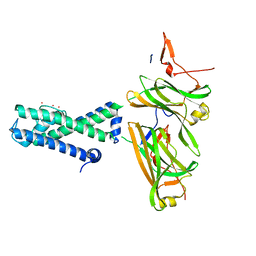 | | POTASSIUM CHANNEL FROM MAGNETOSPIRILLUM MAGNETOTACTICUM | | Descriptor: | POTASSIUM CHANNEL, POTASSIUM ION | | Authors: | Clarke, O.B, Caputo, A.T, Smith, B.J, Gulbis, J.M. | | Deposit date: | 2009-06-24 | | Release date: | 2010-06-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.036 Å) | | Cite: | Domain Reorientation and Rotation of an Intracellular Assembly Regulate Conduction in Kir Potassium Channels.
Cell(Cambridge,Mass.), 141, 2010
|
|
2WLM
 
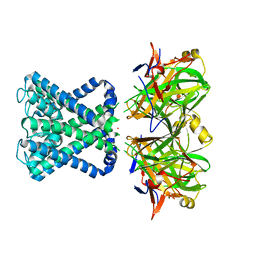 | | POTASSIUM CHANNEL FROM MAGNETOSPIRILLUM MAGNETOTACTICUM | | Descriptor: | POTASSIUM CHANNEL, POTASSIUM ION | | Authors: | Clarke, O.B, Caputo, A.T, Smith, B.J, Gulbis, J.M. | | Deposit date: | 2009-06-24 | | Release date: | 2010-06-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.61 Å) | | Cite: | Domain Reorientation and Rotation of an Intracellular Assembly Regulate Conduction in Kir Potassium Channels.
Cell(Cambridge,Mass.), 141, 2010
|
|
