3U0X
 
 | |
2LUR
 
 | |
3DOJ
 
 | | Structure of Glyoxylate reductase 1 from Arabidopsis (AtGLYR1) | | Descriptor: | CHLORIDE ION, Dehydrogenase-like protein | | Authors: | Jorgensen, R. | | Deposit date: | 2008-07-04 | | Release date: | 2009-07-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Cytosolic NADPH-dependent Glyoxylate Reductase from Arabidopsis: Crystal Structure and Kinetic Characterization of Active Site Mutants
To be Published
|
|
2Q6M
 
 | |
3RX6
 
 | | Crystal structure of Polarity Suppression protein from Enterobacteria phage P4 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, IODIDE ION, MERCURY (II) ION, ... | | Authors: | Banerjee, R, Nath, S, Khamrui, S, Sen, R, Sen, U. | | Deposit date: | 2011-05-10 | | Release date: | 2012-07-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.039 Å) | | Cite: | The first structure of polarity suppression protein, Psu from enterobacteria phage P4, reveals a novel fold and a knotted dimer
J.Biol.Chem., 287, 2012
|
|
5IT2
 
 | | Structure of a transglutaminase 2-specific autoantibody 693-10-B06 Fab fragment | | Descriptor: | heavy chain, light chain | | Authors: | Chen, X, Dalhus, B, Hnida, K, Iversen, R, Sollid, L.M. | | Deposit date: | 2016-03-16 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High abundance of plasma cells secreting transglutaminase 2-specific IgA autoantibodies with limited somatic hypermutation in celiac disease intestinal lesions
Nat. Med., 18, 2012
|
|
2PC0
 
 | | Apo Wild-type HIV Protease in the open conformation | | Descriptor: | MAGNESIUM ION, Protease, R-1,2-PROPANEDIOL | | Authors: | Heaslet, H, Rosenfeld, R, Giffin, M.J, Elder, J.H, McRee, D.E, Stout, C.D. | | Deposit date: | 2007-03-29 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conformational flexibility in the flap domains of ligand-free HIV protease.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
8PM6
 
 | | Human bile salt export pump (BSEP) in complex with inhibitor GBM in nanodiscs | | Descriptor: | 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, Bile salt export pump, CHOLESTEROL | | Authors: | Liu, H, Irobalieva, R.N, Kowal, J, Ni, D, Nosol, K, Bang-Sorensen, R, Lancien, L, Stahlberg, H, Stieger, B, Locher, K.P. | | Deposit date: | 2023-06-28 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural basis of bile salt extrusion and small-molecule inhibition in human BSEP.
Nat Commun, 14, 2023
|
|
8PMJ
 
 | | Vanadate-trapped BSEP in nanodiscs | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Bile salt export pump, ... | | Authors: | Liu, H, Irobalieva, R.N, Kowal, J, Ni, D, Nosol, K, Bang-Sorensen, R, Lancien, L, Stahlberg, H, Stieger, B, Locher, K.P. | | Deposit date: | 2023-06-28 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structural basis of bile salt extrusion and small-molecule inhibition in human BSEP.
Nat Commun, 14, 2023
|
|
8PMD
 
 | | Nucleotide-bound BSEP in nanodiscs | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Bile salt export pump, MAGNESIUM ION | | Authors: | Liu, H, Irobalieva, R.N, Kowal, J, Ni, D, Nosol, K, Bang-Sorensen, R, Lancien, L, Stahlberg, H, Stieger, B, Locher, K.P. | | Deposit date: | 2023-06-28 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis of bile salt extrusion and small-molecule inhibition in human BSEP.
Nat Commun, 14, 2023
|
|
3ZS0
 
 | | Human Myeloperoxidase inactivated by TX2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(4-FLUOROBENZYL)-2-THIOXO-1,2,3,7-TETRAHYDRO-6H-PURIN-6-ONE, ACETATE ION, ... | | Authors: | Tiden, A.K, Sjogren, T, Svensson, M, Bernlind, A, Senthilmohan, R, Auchere, F, Norman, H, Markgren, P.O, Gustavsson, S, Schmidt, S, Lundquist, S, Forbes, L.V, Magon, N.J, Jameson, G.N, Eriksson, H, Kettle, A.J. | | Deposit date: | 2011-06-21 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 2-Thioxanthines are Mechanism-Based Inactivators of Myeloperoxidase that Block Oxidative Stress During Inflammation.
J.Biol.Chem., 286, 2011
|
|
3ZS1
 
 | | Human Myeloperoxidase inactivated by TX5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(2-METHOXYETHYL)-2-THIOXO-1,2,3,7-TETRAHYDRO-6H-PURIN-6-ONE, ACETATE ION, ... | | Authors: | Tiden, A.K, Sjogren, T, Svensson, M, Bernlind, A, Senthilmohan, R, Auchere, F, Norman, H, Markgren, P.O, Gustavsson, S, Schmidt, S, Lundquist, S, Forbes, L.V, Magon, N.J, Jameson, G.N, Eriksson, H, Kettle, A.J. | | Deposit date: | 2011-06-21 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2-Thioxanthines are Mechanism-Based Inactivators of Myeloperoxidase that Block Oxidative Stress During Inflammation.
J.Biol.Chem., 286, 2011
|
|
4V4B
 
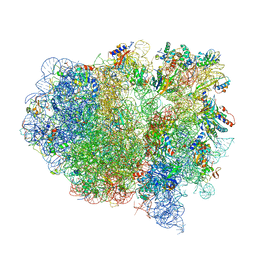 | | Structure of the ribosomal 80S-eEF2-sordarin complex from yeast obtained by docking atomic models for RNA and protein components into a 11.7 A cryo-EM map. | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S11, ... | | Authors: | Spahn, C.M, Gomez-Lorenzo, M.G, Grassucci, R.A, Jorgensen, R, Andersen, G.R, Beckmann, R, Penczek, P.A, Ballesta, J.P.G, Frank, J. | | Deposit date: | 2004-01-06 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (11.7 Å) | | Cite: | Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation.
Embo J., 23, 2004
|
|
7POG
 
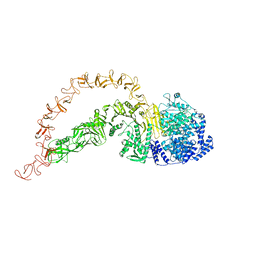 | | High-resolution structure of native toxin A from Clostridioides difficile | | Descriptor: | Toxin A, ZINC ION | | Authors: | Boesen, T, Joergensen, R, Aminzadeh, A, Engelbrecht Larsen, C. | | Deposit date: | 2021-09-08 | | Release date: | 2021-12-08 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | High-resolution structure of native toxin A from Clostridioides difficile.
Embo Rep., 23, 2022
|
|
8RQF
 
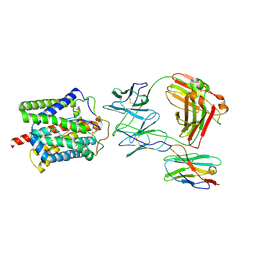 | | Cryo-EM structure of human NTCP-Bulevirtide complex | | Descriptor: | Fab-specific nanobody, Heavy chain of Fab3, Light chain of Fab3, ... | | Authors: | Liu, H, Zakrzewicz, D, Nosol, K, Irobalieva, R.N, Mukherjee, S, Bang-Soerensen, R, Goldmann, N, Kunz, S, Rossi, L, Kossiakoff, A.A, Urban, S, Glebe, D, Geyer, J, Locher, K.P. | | Deposit date: | 2024-01-18 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Cryo-EM structure of human NTCP-Bulevirtide complex
To Be Published
|
|
8DFY
 
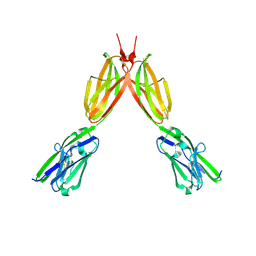 | | Crystal structure of Human BTN2A1 Ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Butyrophilin subfamily 2 member A1, ... | | Authors: | Fulford, T.S, Soliman, C, Castle, R.G, Rigau, M, Ruan, Z, Dolezal, O, Seneviratna, R, Brown, H.G, Hanssen, E, Hammet, A, Li, S, Redmond, S.J, Chung, A, Gorman, M.A, Parker, M.W, Patel, O, Peat, T.S, Newman, J, Behren, A, Gherardin, N.A, Godfrey, D.I, Uldrich, A.P. | | Deposit date: | 2022-06-22 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.552 Å) | | Cite: | Vgamma9-Vdelta2 T cells recognize butyrophilin 2A1 and 3A1 heteromers
To Be Published
|
|
8DFX
 
 | | Crystal structure of Human BTN2A1-BTN3A1 Ectodomain Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Butyrophilin subfamily 2 member A1, Butyrophilin subfamily 3 member A1 | | Authors: | Fulford, T.S, Soliman, C, Castle, R.G, Rigau, M, Ruan, Z, Dolezal, O, Seneviratna, R, Brown, H.G, Hanssen, E, Hammet, A, Li, S, Redmond, S.J, Chung, A, Gorman, M.A, Parker, M.W, Patel, O, Peat, T.S, Newman, J, Behren, A, Gherardin, N.A, Godfrey, D.I, Uldrich, A.P. | | Deposit date: | 2022-06-22 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (5.55 Å) | | Cite: | Vgamma9-Vdelta2 T cells recognize butyrophilin 2A1 and 3A1 heteromers
To Be Published
|
|
8DFW
 
 | | Crystal Structure of Human BTN2A1 in Complex With Vgamma9-Vdelta2 T Cell Receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Butyrophilin subfamily 2 member A1, ... | | Authors: | Fulford, T.S, Soliman, C, Castle, R.G, Rigau, M, Ruan, Z, Dolezal, O, Seneviratna, R, Brown, H.G, Hanssen, E, Hammet, A, Li, S, Redmond, S.J, Chung, A, Gorman, M.A, Parker, M.W, Patel, O, Peat, T.S, Newman, J, Behren, A, Gherardin, N.A, Godfrey, D.I, Uldrich, A.P. | | Deposit date: | 2022-06-22 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Vgamma9-Vdelta2 T cells recognize butyrophilin 2A1 and 3A1 heteromers
To Be Published
|
|
7ZYI
 
 | | Structure of the human sodium/bile acid cotransporter (NTCP) in complex with Fab and nanobody | | Descriptor: | CHOLESTEROL, GLYCOCHENODEOXYCHOLIC ACID, Nanobody, ... | | Authors: | Liu, H, Irobalieva, R.N, Bang-Sorensen, R, Nosol, K, Mukherjee, S, Agrawal, P, Stieger, B, Kossiakoff, A.A, Locher, K.P. | | Deposit date: | 2022-05-24 | | Release date: | 2022-07-13 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structure of human NTCP reveals the basis of recognition and sodium-driven transport of bile salts into the liver.
Cell Res., 32, 2022
|
|
6FJM
 
 | | tubulin-Disorazole Z complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Disorazole Z, ... | | Authors: | Menchon, G, Prota, A.E, Lucena Agell, D, Bucher, P, Jansen, R, Irschik, H, Mueller, R, Paterson, I, Diaz, J.F, Altmann, K.-H, Steinmetz, M.O. | | Deposit date: | 2018-01-22 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A fluorescence anisotropy assay to discover and characterize ligands targeting the maytansine site of tubulin.
Nat Commun, 9, 2018
|
|
7AC1
 
 | | Solution structure of the TAF4-RST domain | | Descriptor: | Transcription initiation factor TFIID subunit 4 | | Authors: | Staby, L, Davidsen, R, Bugge, K, Skriver, K, Kragelund, B.B. | | Deposit date: | 2020-09-09 | | Release date: | 2021-10-06 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | alpha alpha-hub coregulator structure and flexibility determine transcription factor binding and selection in regulatory interactomes.
J.Biol.Chem., 298, 2022
|
|
6Z5N
 
 | | DnaJB1 JD-GF | | Descriptor: | DnaJ homolog subfamily B member 1 | | Authors: | Avraham-Abayev, M, London, N, Rosenzweig, R. | | Deposit date: | 2020-05-27 | | Release date: | 2020-11-18 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | HSP40 proteins use class-specific regulation to drive HSP70 functional diversity.
Nature, 587, 2020
|
|
5F2E
 
 | | Crystal Structure of small molecule ARS-853 covalently bound to K-Ras G12C | | Descriptor: | 1-[3-[4-[2-[[4-chloranyl-5-(1-methylcyclopropyl)-2-oxidanyl-phenyl]amino]ethanoyl]piperazin-1-yl]azetidin-1-yl]prop-2-en-1-one, GLYCEROL, GLYCINE, ... | | Authors: | Patricelli, M.P, Janes, M.R, Li, L.-S, Hansen, R, Peters, U, Kessler, L.V, Chen, Y, Kucharski, J.M, Feng, J, Ely, T, Chen, J.H, Firdaus, S.J, Babbar, A, Ren, P, Liu, Y. | | Deposit date: | 2015-12-01 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Selective Inhibition of Oncogenic KRAS Output with Small Molecules Targeting the Inactive State.
Cancer Discov, 6, 2016
|
|
6FXB
 
 | | Bovine beta-lactoglobulin variant A at pH 4.0 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Major allergen beta-lactoglobulin, NITRATE ION | | Authors: | Khan, S, Ipsen, R, Almdal, K, Svensson, B, Harris, P. | | Deposit date: | 2018-03-08 | | Release date: | 2018-05-23 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Revealing the Dimeric Crystal and Solution Structure of beta-Lactoglobulin at pH 4 and Its pH and Salt Dependent Monomer-Dimer Equilibrium.
Biomacromolecules, 19, 2018
|
|
4ZD3
 
 | | Structure of a transglutaminase 2-specific autoantibody Fab fragment | | Descriptor: | 679-14-14E06 Fab fragment heavy chain, 679-14-14E06 Fab fragment light chain | | Authors: | Chen, X, Dalhus, B, Hnida, K, Iversen, R, Sollid, L.M. | | Deposit date: | 2015-04-16 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Antigen Recognition by Transglutaminase 2-specific Autoantibodies in Celiac Disease.
J.Biol.Chem., 290, 2015
|
|
