3JCX
 
 | | Canine Parvovirus complexed with Fab E | | Descriptor: | Capsid protein 2, Fab E heavy chain, Fab E light chain | | Authors: | Organtini, L.J, Iketani, S, Huang, K, Ashley, R.E, Makhov, A.M, Conway, J.F, Parrish, C.R, Hafenstein, S. | | Deposit date: | 2016-03-21 | | Release date: | 2016-07-20 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Near-Atomic Resolution Structure of a Highly Neutralizing Fab Bound to Canine Parvovirus.
J.Virol., 90, 2016
|
|
3JSD
 
 | | Insulin's biosynthesis and activity have opposing structural requirements: a new factor in neonatal diabetes mellitus | | Descriptor: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | Authors: | Weiss, M.A, Wan, Z.L, Dodson, E.J, Liu, M, Xu, B, Hua, Q.X, Turkenburg, M, Whittingham, J, Nakagawa, S.H, Huang, K, Hu, S.Q, Jia, W.H, Wang, S.H, Brange, J, Whittaker, J, Arvan, P, Katsoyannis, P.G, Dodson, G.G. | | Deposit date: | 2009-09-10 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insulin's biosynthesis and activity have opposing structural requirements: a new factor in neonatal diabetes mellitus
To be Published
|
|
1XF7
 
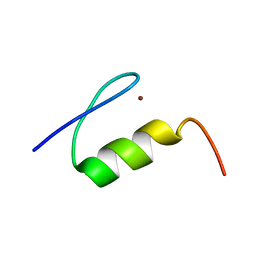 | | High Resolution NMR Structure of the Wilms' Tumor Suppressor Protein (WT1) Finger 3 | | Descriptor: | Wilms' Tumor Protein, ZINC ION | | Authors: | Lachenmann, M.J, Ladbury, J.E, Dong, J, Huang, K, Carey, P, Weiss, M.A. | | Deposit date: | 2004-09-14 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Why zinc fingers prefer zinc: ligand-field symmetry and the hidden thermodynamics of metal ion selectivity
Biochemistry, 43, 2004
|
|
1XW7
 
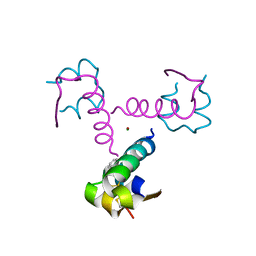 | | Diabetes-Associated Mutations in Human Insulin: Crystal Structure and Photo-Cross-Linking Studies of A-Chain Variant Insulin Wakayama | | Descriptor: | CHLORIDE ION, Insulin, PHENOL, ... | | Authors: | Wan, Z.L, Huang, K, Xu, B, Chu, Y.C, Hu, S.Q, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2004-10-29 | | Release date: | 2005-04-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Diabetes-associated mutations in human insulin: crystal structure and photo-cross-linking studies of a-chain variant insulin wakayama
Biochemistry, 44, 2005
|
|
1XRZ
 
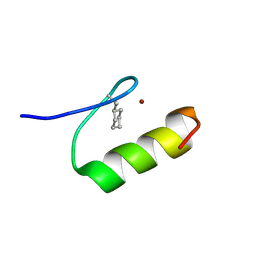 | | NMR Structure of a Zinc Finger with Cyclohexanylalanine Substituted for the Central Aromatic Residue | | Descriptor: | ZINC ION, Zinc finger Y-chromosomal protein | | Authors: | Lachenmann, M.J, Ladbury, J.E, Qian, X, Huang, K, Singh, R, Weiss, M.A. | | Deposit date: | 2004-10-17 | | Release date: | 2004-11-30 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Solvation and the hidden thermodynamics of a zinc finger probed
by nonstandard repair of a protein crevice
Protein Sci., 13, 2004
|
|
6TEP
 
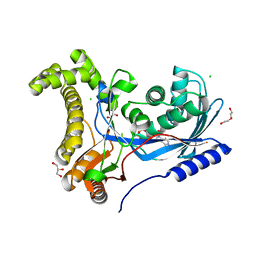 | | Crystal structure of a galactokinase from Bifidobacterium infantis in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Keenan, T, Parmeggiani, F, Fontenelle, C.Q, Malassis, J, Vendeville, J, Offen, W.A, Both, P, Huang, K, Marchesi, A, Heyam, A, Young, C, Charnock, S, Davies, G.J, Linclau, B, Flitsch, S.L, Fascione, M.A. | | Deposit date: | 2019-11-12 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Profiling Substrate Promiscuity of Wild-Type Sugar Kinases for Multi-fluorinated Monosaccharides.
Cell Chem Biol, 27, 2020
|
|
1DBU
 
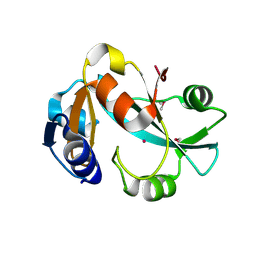 | | Crystal structure of cysteinyl-tRNA(Pro) deacylase protein from H. influenzae (HI1434) | | Descriptor: | MERCURY (II) ION, cysteinyl-tRNA(Pro) deacylase | | Authors: | Zhang, H, Huang, K, Li, Z, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 1999-11-03 | | Release date: | 2000-06-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of YbaK protein from Haemophilus influenzae (HI1434) at 1.8 A resolution: functional implications.
Proteins, 40, 2000
|
|
1DS3
 
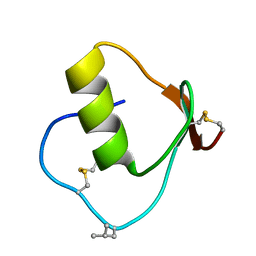 | | CRYSTAL STRUCTURE OF OMTKY3-CH2-ASP19I | | Descriptor: | OVOMUCOID | | Authors: | Bateman, K.S, Huang, K, Anderson, S, Lu, W, Qasim, M.A, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 2000-01-06 | | Release date: | 2001-01-31 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Contribution of peptide bonds to inhibitor-protease binding: crystal structures of the turkey ovomucoid third domain backbone variants OMTKY3-Pro18I and OMTKY3-psi[COO]-Leu18I in complex with Streptomyces griseus proteinase B (SGPB) and the structure of the free inhibitor, OMTKY-3-psi[CH2NH2+]-Asp19I
J.Mol.Biol., 305, 2001
|
|
6TEQ
 
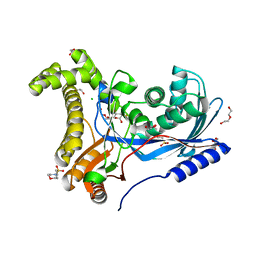 | | Crystal structure of a galactokinase from Bifidobacterium infantis in complex with 2-deoxy-2-fluoro-galactose | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-deoxy-2-fluoro-alpha-D-galactopyranose, ... | | Authors: | Keenan, T, Parmeggiani, F, Fontenelle, C.Q, Malassis, J, Vendeville, J, Offen, W.A, Both, P, Huang, K, Marchesi, A, Heyam, A, Young, C, Charnock, S, Davies, G.J, Linclau, B, Flitsch, S.L, Fascione, M.A. | | Deposit date: | 2019-11-12 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Profiling Substrate Promiscuity of Wild-Type Sugar Kinases for Multi-fluorinated Monosaccharides.
Cell Chem Biol, 27, 2020
|
|
6TER
 
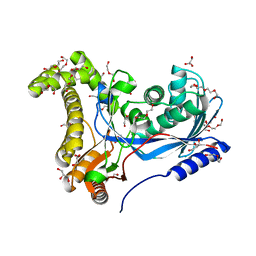 | | Crystal structure of a galactokinase from Bifidobacterium infantis in complex with Galactose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Keenan, T, Parmeggiani, F, Fontenelle, C.Q, Malassis, J, Vendeville, J, Offen, W.A, Both, P, Huang, K, Marchesi, A, Heyam, A, Young, C, Charnock, S, Davies, G.J, Linclau, B, Flitsch, S.L, Fascione, M.A. | | Deposit date: | 2019-11-12 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Profiling Substrate Promiscuity of Wild-Type Sugar Kinases for Multi-fluorinated Monosaccharides.
Cell Chem Biol, 27, 2020
|
|
1DS2
 
 | | CRYSTAL STRUCTURE OF SGPB:OMTKY3-COO-LEU18I | | Descriptor: | OVOMUCOID, PROTEINASE B (SGPB) | | Authors: | Bateman, K.S, Huang, K, Anderson, S, Lu, W, Qasim, M.A, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 2000-01-06 | | Release date: | 2001-01-31 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Contribution of peptide bonds to inhibitor-protease binding: crystal structures of the turkey ovomucoid third domain backbone variants OMTKY3-Pro18I and OMTKY3-psi[COO]-Leu18I in complex with Streptomyces griseus proteinase B (SGPB) and the structure of the free inhibitor, OMTKY-3-psi[CH2NH2+]-Asp19I
J.Mol.Biol., 305, 2001
|
|
1DBX
 
 | | Crystal structure of cysteinyl-tRNA(Pro) deacylase from H. influenzae (HI1434) | | Descriptor: | cysteinyl-tRNA(Pro) deacylase | | Authors: | Zhang, H, Huang, K, Li, Z, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 1999-11-03 | | Release date: | 2000-06-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of YbaK protein from Haemophilus influenzae (HI1434) at 1.8 A resolution: functional implications.
Proteins, 40, 2000
|
|
2JZQ
 
 | | Design of an Active Ultra-Stable Single-Chain Insulin Analog 20 Structures | | Descriptor: | Insulin | | Authors: | Hua, Q.X, Nakarawa, S, Jia, W.H, Huang, K, Philips, N.F, Hu, S.Q, Weiss, M.A. | | Deposit date: | 2008-01-11 | | Release date: | 2008-02-26 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Design of an active ultrastable single-chain insulin analog: synthesis, structure, and therapeutic implications.
J.Biol.Chem., 283, 2008
|
|
3V19
 
 | | Forestalling insulin fibrillation by insertion of a chiral clamp mechanism-based application of protein engineering to global health | | Descriptor: | CHLORIDE ION, Insulin, PHENOL, ... | | Authors: | Wan, Z.L, Hua, Q.X, Wickramasinghe, N.P, Huang, K, Petkova, A.T, Hu, S.Q, Phillips, N.B, Yeh, I.J, Whittake, J, Ismail-Beigi, F, Katsoyyannis, P.G, Tycko, R, Weiss, M.A. | | Deposit date: | 2011-12-09 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Forestalling insulin fibrillation by insertion of a
chiral clamp mechanism-based application of protein engineering to global health
To be Published
|
|
3V1G
 
 | | Forestalling insulin fibrillation by insertion of a chiral clamp mechanism-based application of protein engineering to global health | | Descriptor: | CHLORIDE ION, Insulin, PHENOL, ... | | Authors: | Wan, Z.L, Hua, Q.X, Wickramasinghe, N.P, Huang, K, Petkova, A.T, Hu, S.Q, Phillips, N.B, Yeh, I.J, Whittake, J, Ismail-Beigi, F, Katsoyyannis, P.G, Tycko, R, Weiss, M.A. | | Deposit date: | 2011-12-09 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Forestalling insulin fibrillation by insertion of a
chiral clamp mechanism-based application of protein engineering to global health
To be Published
|
|
6BP2
 
 | | Therapeutic human monoclonal antibody MR191 bound to a marburgvirus glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein, Envelope glycoprotein GP2, ... | | Authors: | King, L.B, Fusco, M.L, Flyak, A.I, Ilinykh, P.A, Huang, K, Gunn, B, Kirchdoerfer, R.N, Hastie, K.M, Sangha, A.K, Meiler, J, Alter, G, Bukreyev, A, Crowe, J.E.J, Saphire, E.O. | | Deposit date: | 2017-11-21 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.172 Å) | | Cite: | The Marburgvirus-Neutralizing Human Monoclonal Antibody MR191 Targets a Conserved Site to Block Virus Receptor Binding.
Cell Host Microbe, 23, 2018
|
|
2L1Y
 
 | | NMR Structure of human insulin mutant GLY-B20-D-ALA, GLY-B23-D-ALA PRO-B28-LYS, LYS-B29-PRO, 20 Structures | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Wan, Z.L, Hua, Q.X, Huang, K, Hu, S.Q, Philips, N.B, Katsoyannis, J.W, Weiss, M.A. | | Deposit date: | 2010-08-09 | | Release date: | 2011-08-31 | | Method: | SOLUTION NMR | | Cite: | Chiral Protein Engineering and its Application in G Health
To be Published
|
|
2L1Z
 
 | | NMR Structure of human insulin mutant GLY-B20-D-ALA, GLY-B23-D-ALA PRO-B28-LYS, LYS-B29-PRO, 20 Structures | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Wan, Z.L, Hua, Q.X, Huang, K, Hu, S.Q, Philips, N.B, Katsoyannis, J.W, Weiss, M.A. | | Deposit date: | 2010-08-09 | | Release date: | 2011-08-31 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | Chiral Protein Engineering and its Application in G Health
To be Published
|
|
2NU0
 
 | | Molecular structures of the complexes of SGPB with OMTKY3 aromatic P1 variants Trp18I, His18I, Phe18I, and Tyr18I | | Descriptor: | Ovomucoid, Streptogrisin B, Protease B | | Authors: | Bateman, K.S, Anderson, S, Lu, W, Qasim, M.A, Huang, K, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 2006-11-08 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular structures of the complexes of SGPB with OMTKY3 aromatic P1 variants Trp18I, His18I, Phe18I, and Tyr18I
To be Published
|
|
2NU1
 
 | | Molecular structures of the complexes of SGPB with OMTKY3 aromatic P1 variants Trp18I, His18I, Phe18I and Tyr18I | | Descriptor: | Ovomucoid, Streptogrisin B | | Authors: | Bateman, K.S, Anderson, S, Lu, W, Qasim, M.A, Huang, K, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 2006-11-08 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular structures of the complexes of SGPB with OMTKY3 aromatic P1 variants Trp18I, His18I, Phe18I and Tyr18I
To be Published
|
|
2NU3
 
 | | Accommodation of positively-charged residues in a hydrophobic specificity pocket: Crystal structures of SGPB in complex with OMTKY3 variants Lys18I and Arg18I | | Descriptor: | Ovomucoid, Streptogrisin B, Proteinase B | | Authors: | Bateman, K.S, Anderson, S, Lu, W, Qasim, M.A, Huang, K, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 2006-11-08 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Accommodation of positively-charged residues in a hydrophobic specificity pocket: Crystal structures of SGPB in complex with OMTKY3 variants Lys18I and Arg18I
To be Published
|
|
3R8F
 
 | | Replication initiator DnaA bound to AMPPCP and single-stranded DNA | | Descriptor: | 5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3', Chromosomal replication initiator protein dnaA, MAGNESIUM ION, ... | | Authors: | Duderstadt, K.E, Chuang, K, Berger, J.M. | | Deposit date: | 2011-03-23 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.366 Å) | | Cite: | DNA stretching by bacterial initiators promotes replication origin opening.
Nature, 478, 2011
|
|
2JSF
 
 | |
8HZM
 
 | | A new fluorescent RNA aptamer bound with N, manganese soak | | Descriptor: | (5~{Z})-5-[[4-[2-hydroxyethyl(methyl)amino]phenyl]methylidene]-3-methyl-2-[(~{E})-2-phenylethenyl]imidazol-4-one, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2023-01-09 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of a small monomeric Clivia fluorogenic RNA with a large Stokes shift.
Nat.Chem.Biol., 2024
|
|
8HZE
 
 | | A new fluorescent RNA aptamer bound with N | | Descriptor: | (5~{Z})-5-[[4-[2-hydroxyethyl(methyl)amino]phenyl]methylidene]-3-methyl-2-[(~{E})-2-phenylethenyl]imidazol-4-one, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2023-01-09 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural basis of a small monomeric Clivia fluorogenic RNA with a large Stokes shift.
Nat.Chem.Biol., 2024
|
|
