3E2E
 
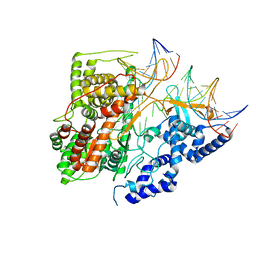 | | Crystal Structure of an Intermediate Complex of T7 RNAP and 7nt of RNA | | Descriptor: | DNA (28-MER), DNA (31-MER), DNA-directed RNA polymerase, ... | | Authors: | Durniak, K.J, Bailey, S, Steitz, T.A. | | Deposit date: | 2008-08-05 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of a transcribing t7 RNA polymerase in transition from initiation to elongation
Science, 322, 2008
|
|
3EDU
 
 | | Crystal structure of the ankyrin-binding domain of human erythroid spectrin | | Descriptor: | Spectrin beta chain, erythrocyte | | Authors: | Simonovic, M, Stabach, P, Simonovic, I, Steitz, T.A, Morrow, J.S. | | Deposit date: | 2008-09-03 | | Release date: | 2009-02-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of the ankyrin-binding site of {beta}-spectrin reveals how tandem spectrin-repeats generate unique ligand-binding properties
Blood, 113, 2009
|
|
4V8I
 
 | | Crystal structure of YfiA bound to the 70S ribosome. | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Polikanov, Y.S, Blaha, G.M, Steitz, T.A. | | Deposit date: | 2011-12-12 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | How hibernation factors RMF, HPF, and YfiA turn off protein synthesis.
Science, 336, 2012
|
|
4V9R
 
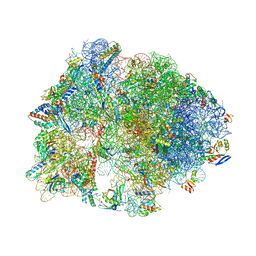 | | Crystal structure of antibiotic DITYROMYCIN bound to 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Bulkley, D.P, Brandi, L, Polikanov, Y.S, Fabbretti, A, O'Connor, M, Gualerzi, C.O, Steitz, T.A. | | Deposit date: | 2013-12-05 | | Release date: | 2014-07-09 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The antibiotics dityromycin and GE82832 bind protein S12 and block EF-G-catalyzed translocation.
Cell Rep, 6, 2014
|
|
4V7Y
 
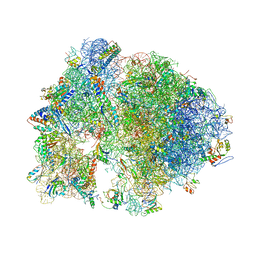 | | Structure of the Thermus thermophilus 70S ribosome complexed with azithromycin. | | Descriptor: | 16S rRNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Bulkley, D.P, Innis, C.A, Blaha, G, Steitz, T.A. | | Deposit date: | 2010-08-18 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Revisiting the structures of several antibiotics bound to the bacterial ribosome.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4V8H
 
 | | Crystal structure of HPF bound to the 70S ribosome. | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Polikanov, Y.S, Blaha, G.M, Steitz, T.A. | | Deposit date: | 2011-12-11 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | How Hibernation Factors RMF, HPF, and YfiA Turn Off Protein Synthesis.
Science, 336, 2012
|
|
4V7X
 
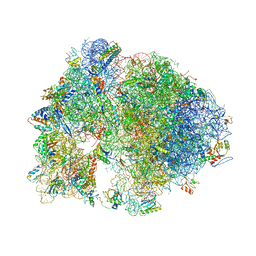 | | Structure of the Thermus thermophilus ribosome complexed with erythromycin. | | Descriptor: | 16S rRNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Bulkley, D.P, Innis, C.A, Blaha, G, Steitz, T.A. | | Deposit date: | 2010-08-17 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Revisiting the structures of several antibiotics bound to the bacterial ribosome.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4V9S
 
 | | Crystal structure of antibiotic GE82832 bound to 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Bulkley, D.P, Brandi, L, Polikanov, Y.S, Fabbretti, A, O'Connor, M, Gualerzi, C.O, Steitz, T.A. | | Deposit date: | 2013-12-05 | | Release date: | 2014-07-09 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The antibiotics dityromycin and GE82832 bind protein S12 and block EF-G-catalyzed translocation.
Cell Rep, 6, 2014
|
|
4V95
 
 | | Crystal structure of YAEJ bound to the 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Gagnon, M.G, Seetharaman, S.V, Bulkley, D.P, Steitz, T.A. | | Deposit date: | 2012-01-27 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the rescue of stalled ribosomes: structure of YaeJ bound to the ribosome.
Science, 335, 2012
|
|
4V8A
 
 | | The structure of thermorubin in complex with the 70S ribosome from Thermus thermophilus. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Bulkley, D, Johnson, F.A, Steitz, T.A. | | Deposit date: | 2011-12-05 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The antibiotic thermorubin inhibits protein synthesis by binding to inter-subunit bridge b2a of the ribosome.
J.Mol.Biol., 416, 2012
|
|
1QSL
 
 | |
1REA
 
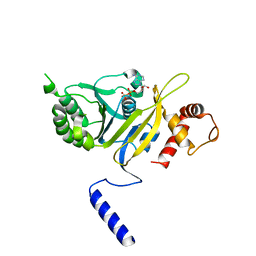 | |
5DOX
 
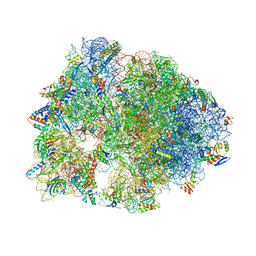 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with Hygromycin-A at 3.1A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Polikanov, Y.S, Starosta, A.L, Juette, M.F, Altman, R.B, Terry, D.S, Lu, W, Burnett, B.J, Dinos, G, Reynolds, K, Blanchard, S.C, Steitz, T.A, Wilson, D.N. | | Deposit date: | 2015-09-11 | | Release date: | 2015-12-30 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Distinct tRNA Accommodation Intermediates Observed on the Ribosome with the Antibiotics Hygromycin A and A201A.
Mol.Cell, 58, 2015
|
|
5DOY
 
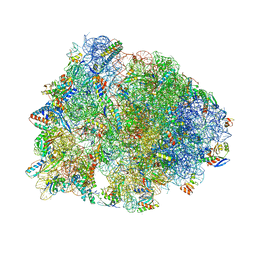 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with antibiotic Hygromycin A, mRNA and three tRNAs in the A, P and E sites at 2.6A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Polikanov, Y.S, Starosta, A.L, Juette, M.F, Altman, R.B, Terry, D.S, Lu, W, Burnett, B.J, Dinos, G, Reynolds, K, Blanchard, S.C, Steitz, T.A, Wilson, D.N. | | Deposit date: | 2015-09-11 | | Release date: | 2015-12-30 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Distinct tRNA Accommodation Intermediates Observed on the Ribosome with the Antibiotics Hygromycin A and A201A.
Mol.Cell, 58, 2015
|
|
1TV6
 
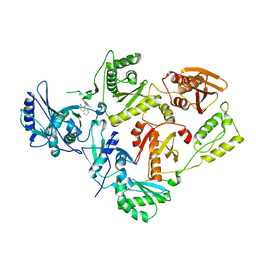 | | HIV-1 Reverse Transcriptase Complexed with CP-94,707 | | Descriptor: | 3-[4-(2-METHYL-IMIDAZO[4,5-C]PYRIDIN-1-YL)BENZYL]-3H-BENZOTHIAZOL-2-ONE, reverse transcriptase p51 subunit, reverse transcriptase p66 subunit | | Authors: | Pata, J.D, Stirtan, W.G, Goldstein, S.W, Steitz, T.A. | | Deposit date: | 2004-06-28 | | Release date: | 2004-07-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of HIV-1 reverse transcriptase bound to an inhibitor active against mutant RTs resistant to other non-nucleoside inhibitors
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
6MS4
 
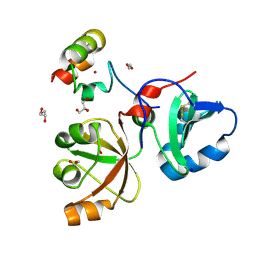 | | Crystal structure of the DENR-MCT-1 complex | | Descriptor: | Density-regulated protein, GLYCEROL, Malignant T-cell-amplified sequence 1, ... | | Authors: | Lomakin, I.B, Steitz, T.A, Dmitriev, S.E. | | Deposit date: | 2018-10-16 | | Release date: | 2019-01-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal structure of the DENR-MCT-1 complex revealed zinc-binding site essential for heterodimer formation.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
1VQ7
 
 | |
1VQN
 
 | |
1VQ6
 
 | |
1XI1
 
 | | Phi29 DNA polymerase ssDNA complex, monoclinic crystal form | | Descriptor: | 5'-D(P*TP*TP*TP*TP*T)-3', DNA polymerase, MAGNESIUM ION | | Authors: | Kamtekar, S, Berman, A.J, Wang, J, Lazaro, J.M, de Vega, M, Blanco, L, Salas, M, Steitz, T.A. | | Deposit date: | 2004-09-21 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Correction of X-ray intensities from single crystals containing lattice-translocation defects
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1XHX
 
 | | Phi29 DNA Polymerase, orthorhombic crystal form | | Descriptor: | DNA polymerase, MAGNESIUM ION, SULFATE ION | | Authors: | Kamtekar, S, Berman, A.J, Wang, J, Lazaro, J.M, de Vega, M, Blanco, L, Salas, M, Steitz, T.A. | | Deposit date: | 2004-09-21 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Insights into Strand Displacement and Processivity from the Crystal Structure of the Protein-Primed DNA Polymerase of Bacteriophage phi29
Mol.Cell, 16, 2004
|
|
1XHZ
 
 | | Phi29 DNA polymerase, orthorhombic crystal form, ssDNA complex | | Descriptor: | 5'-D(*TP*TP*TP*TP*T)-3', DNA polymerase | | Authors: | Kamtekar, S, Berman, A.J, Wang, J, Lazaro, J.M, de Vega, M, Blanco, L, Salas, M, Steitz, T.A. | | Deposit date: | 2004-09-21 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights into Strand Displacement and Processivity from the Crystal Structure of the Protein-Primed DNA Polymerase of Bacteriophage phi29
Mol.Cell, 16, 2004
|
|
1B8H
 
 | | SLIDING CLAMP, DNA POLYMERASE | | Descriptor: | DNA POLYMERASE PROCESSIVITY COMPONENT, DNA POLYMERASE fragment | | Authors: | Shamoo, Y, Steitz, T.A. | | Deposit date: | 1999-02-01 | | Release date: | 1999-02-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Building a replisome from interacting pieces: sliding clamp complexed to a peptide from DNA polymerase and a polymerase editing complex.
Cell(Cambridge,Mass.), 99, 1999
|
|
1BDN
 
 | |
1B77
 
 | |
