2IT7
 
 | |
6LQI
 
 | | Cryo-EM structure of the mouse Piezo1 isoform Piezo1.1 | | Descriptor: | Piezo-type mechanosensitive ion channel component 1 | | Authors: | Geng, J, Liu, W, Zhou, H, Zhang, T, Wang, L, Zhang, M, Shen, B, Li, X, Xiao, B. | | Deposit date: | 2020-01-13 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | A Plug-and-Latch Mechanism for Gating the Mechanosensitive Piezo Channel.
Neuron, 106, 2020
|
|
7VPU
 
 | |
2ZKA
 
 | | Urate oxidase complexed with 8-azaxanthine under 1.0 MPa oxygen pressure | | Descriptor: | 8-AZAXANTHINE, OXYGEN MOLECULE, SODIUM ION, ... | | Authors: | Colloc'h, N, Gabison, L, Chiadmi, M, Abraini, J.H, Prange, T. | | Deposit date: | 2008-03-13 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Oxygen pressurized X-ray crystallography: probing the dioxygen binding site in cofactorless urate oxidase and implications for its catalytic mechanism.
Biophys.J., 95, 2008
|
|
6M9Z
 
 | |
7ZGM
 
 | | Plant N-glycan specific alpha-1,3-mannosidase | | Descriptor: | CALCIUM ION, Glycoside hydrolase family 92 protein, POTASSIUM ION, ... | | Authors: | Basle, A, Crouch, L, Bolam, D. | | Deposit date: | 2022-04-04 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Plant N -glycan breakdown by human gut Bacteroides.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7ZGN
 
 | |
6MCT
 
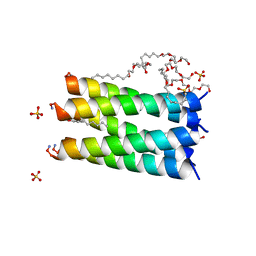 | |
6ME2
 
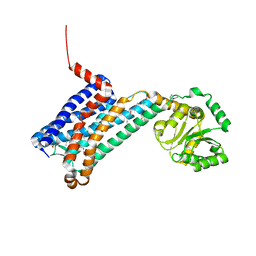 | | XFEL crystal structure of human melatonin receptor MT1 in complex with ramelteon | | Descriptor: | DI(HYDROXYETHYL)ETHER, N-{2-[(8S)-1,6,7,8-tetrahydro-2H-indeno[5,4-b]furan-8-yl]ethyl}propanamide, OLEIC ACID, ... | | Authors: | Stauch, B, Johansson, L.C, McCorvy, J.D, Patel, N, Han, G.W, Gati, C, Batyuk, A, Ishchenko, A, Brehm, W, White, T.A, Michaelian, N, Madsen, C, Zhu, L, Grant, T.D, Grandner, J.M, Olsen, R.H.J, Tribo, A.R, Weierstall, U, Roth, B.L, Katritch, V, Liu, W, Cherezov, V. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of ligand recognition at the human MT1melatonin receptor.
Nature, 569, 2019
|
|
1SKT
 
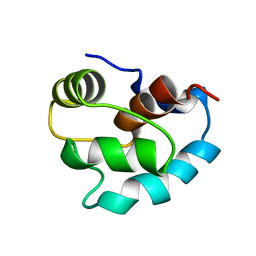 | | SOLUTION STRUCTURE OF APO N-DOMAIN OF TROPONIN C, NMR, 40 STRUCTURES | | Descriptor: | TROPONIN-C | | Authors: | Tsuda, S, Miura, A, Gagne, S.M, Spyracopoulos, L, Sykes, B.D. | | Deposit date: | 1998-04-06 | | Release date: | 1998-12-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Low-temperature-induced structural changes in the Apo regulatory domain of skeletal muscle troponin C.
Biochemistry, 38, 1999
|
|
2ZAV
 
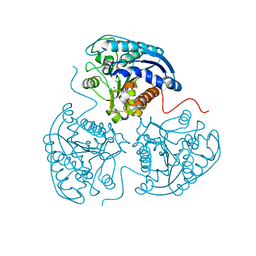 | |
2ZCI
 
 | |
6LYP
 
 | | Cryo-EM structure of AtMSL1 wild type | | Descriptor: | Mechanosensitive ion channel protein 1, mitochondrial | | Authors: | Sun, L. | | Deposit date: | 2020-02-15 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural Insights into a Plant Mechanosensitive Ion Channel MSL1.
Cell Rep, 30, 2020
|
|
7ZJP
 
 | | Optimization of TEAD P-Site Binding Fragment Hit into In Vivo Active Lead MSC-4106 | | Descriptor: | 2-methyl-4-[4-(trifluoromethyl)phenyl]pyrazolo[3,4-b]indole-7-carboxylic acid, SULFATE ION, Transcriptional enhancer factor TEF-1 | | Authors: | Freire, F, Heinrich, T, Petersson, C, Schneider, R, Garg, S, Schwarz, D, Gunera, J, Seshire, A, Koetzner, L, Schlesiger, S, Musil, D, Schilke, H, Doerfel, B, Diehl, P, Boepple, P, Lemos, A.R, Sousa, P.M.F, Freire, F, Bandeiras, T.M, Carswell, E, Pearson, N, Sirohi, S, Hooker, M, Trivier, E, Broome, R, Balsiger, A, Crowden, A, Dillon, C, Wienke, D. | | Deposit date: | 2022-04-11 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Optimization of TEAD P-Site Binding Fragment Hit into In Vivo Active Lead MSC-4106 .
J.Med.Chem., 65, 2022
|
|
6ME4
 
 | | XFEL crystal structure of human melatonin receptor MT1 in complex with 2-iodomelatonin | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, N-[2-(2-iodo-5-methoxy-1H-indol-3-yl)ethyl]acetamide, ... | | Authors: | Stauch, B, Johansson, L.C, McCorvy, J.D, Patel, N, Han, G.W, Gati, C, Batyuk, A, Ishchenko, A, Brehm, W, White, T.A, Michaelian, N, Madsen, C, Zhu, L, Grant, T.D, Grandner, J.M, Olsen, R.H.J, Tribo, A.R, Weierstall, U, Roth, B.L, Katritch, V, Liu, W, Cherezov, V. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of ligand recognition at the human MT1melatonin receptor.
Nature, 569, 2019
|
|
1R6B
 
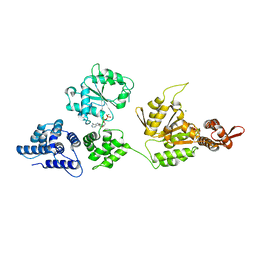 | | High resolution crystal structure of ClpA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ClpA protein, MAGNESIUM ION | | Authors: | Xia, D, Maurizi, M.R, Guo, F, Singh, S.K, Esser, L. | | Deposit date: | 2003-10-15 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystallographic investigation of peptide binding sites in the N-domain of the ClpA chaperone.
J.Struct.Biol., 146, 2004
|
|
6LO8
 
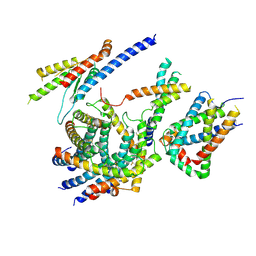 | | Cryo-EM structure of the TIM22 complex from yeast | | Descriptor: | Mitochondrial import inner membrane translocase subunit TIM10, Mitochondrial import inner membrane translocase subunit TIM12, Mitochondrial import inner membrane translocase subunit TIM18, ... | | Authors: | Zhang, Y, Zhou, X, Wu, X, Li, L. | | Deposit date: | 2020-01-04 | | Release date: | 2020-09-30 | | Last modified: | 2021-03-17 | | Method: | ELECTRON MICROSCOPY (3.83 Å) | | Cite: | Structure of the mitochondrial TIM22 complex from yeast.
Cell Res., 31, 2021
|
|
3A3W
 
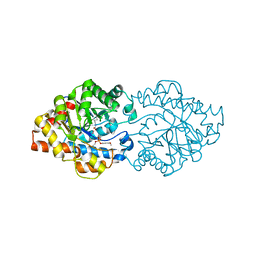 | | Structure of OpdA mutant (G60A/A80V/S92A/R118Q/K185R/Q206P/D208G/I260T/G273S) with diethyl 4-methoxyphenyl phosphate bound in the active site | | Descriptor: | COBALT (II) ION, DIETHYL 4-METHOXYPHENYL PHOSPHATE, Phosphotriesterase | | Authors: | Ollis, D.L, Tawfik, D.S, Schenk, G, Jackson, C.J, Foo, J.L, Tokuriki, N, Afriat, L, Carr, P.D, Kim, H.K. | | Deposit date: | 2009-06-23 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conformational sampling, catalysis, and evolution of the bacterial phosphotriesterase
Proc.Natl.Acad.Sci.USA, 2009
|
|
6M79
 
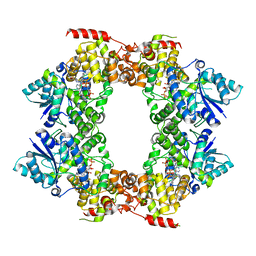 | |
2HQV
 
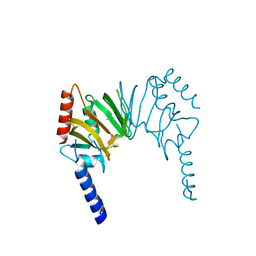 | | X-ray Crystal Structure of Protein AGR_C_4470 from Agrobacterium tumefaciens. Northeast Structural Genomics Consortium Target AtR92. | | Descriptor: | AGR_C_4470p | | Authors: | Vorobiev, S.M, Neely, H, Seetharaman, J, Zhao, L, Cunningham, K, Ma, L.C, Fang, Y, Xiao, R, Acton, T, Montelione, T.G, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-07-19 | | Release date: | 2006-09-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of AGR_C_4470p from Agrobacterium tumefaciens.
Protein Sci., 16, 2007
|
|
6MFT
 
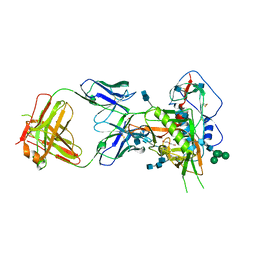 | | Crystal structure of glycosylated 426c HIV-1 gp120 core G459C in complex with glVRC01 A60C heavy chain | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Weidle, C, Pancera, M, Stamatatos, L, Gray, M. | | Deposit date: | 2018-09-12 | | Release date: | 2018-11-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.315 Å) | | Cite: | Germline VRC01 antibody recognition of a modified clade C HIV-1 envelope trimer and a glycosylated HIV-1 gp120 core.
Elife, 7, 2018
|
|
1RZW
 
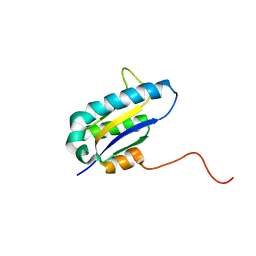 | | The Solution Structure of the Archaeglobus fulgidis protein AF2095. Northeast Structural Genomics Consortium target GR4 | | Descriptor: | Protein AF2095(GR4) | | Authors: | Powers, R, Acton, T.B, Huang, Y.J, Liu, J, Ma, L, Rost, B, Chiang, Y, Cort, J.R, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-12-29 | | Release date: | 2004-11-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Archaeglobus fulgidis peptidyl-tRNA hydrolase (Pth2) provides evidence for an extensive conserved family of Pth2 enzymes in archea, bacteria, and eukaryotes
Protein Sci., 14, 2005
|
|
7L0R
 
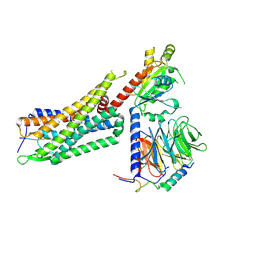 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, noncanonical state, without AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
1S55
 
 | | Mouse RANKL Structure at 1.9A Resolution | | Descriptor: | CHLORIDE ION, Tumor necrosis factor ligand superfamily member 11 | | Authors: | Teale, M.J, Feug, X, Chen, L, Bice, T, Meehan, E.J. | | Deposit date: | 2004-01-19 | | Release date: | 2005-07-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Murine RANKL Extra Cellular Domain Homotrimer Structure In Space Groups P212121 And H3 At 1.9 And 2.6 Respectively
To be Published
|
|
6M54
 
 | | Human apo ferritin frozen on TEM grid with Amorphous nickel titanium alloy supporting film | | Descriptor: | FE (II) ION, Ferritin heavy chain | | Authors: | Huang, X, Zhang, L, Wen, Z, Chen, H, Li, S, Ji, G, Yin, C, Sun, F. | | Deposit date: | 2020-03-09 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Amorphous nickel titanium alloy film: A new choice for cryo electron microscopy sample preparation.
Prog.Biophys.Mol.Biol., 156, 2020
|
|
