9AX6
 
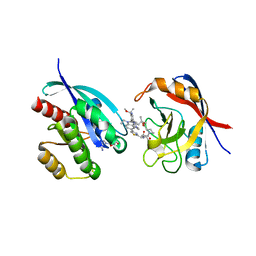 | | Tricomplex of RMC-6236, KRAS G12D, and CypA | | Descriptor: | (1R,2S)-N-[(1P,7S,9S,13R,20M)-21-ethyl-20-{2-[(1R)-1-methoxyethyl]-5-(4-methylpiperazin-1-yl)pyridin-3-yl}-17,17-dimethyl-8,14-dioxo-15-oxa-4-thia-9,21,27,28-tetraazapentacyclo[17.5.2.1~2,5~.1~9,13~.0~22,26~]octacosa-1(24),2,5(28),19,22,25-hexaen-7-yl]-2-methylcyclopropane-1-carboxamide, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Tomlinson, A.C.A, Saldajeno-Concar, M, Knox, J.E, Yano, J.K. | | Deposit date: | 2024-03-05 | | Release date: | 2024-04-17 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Translational and Therapeutic Evaluation of RAS-GTP Inhibition by RMC-6236 in RAS-Driven Cancers.
Cancer Discov, 14, 2024
|
|
8JXS
 
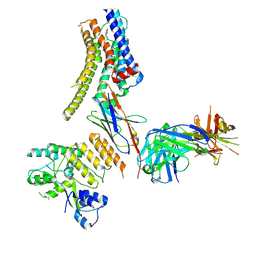 | | Structure of nanobody-bound DRD1_PF-6142 complex | | Descriptor: | 4-[3-methyl-4-(6-methylimidazo[1,2-a]pyrazin-5-yl)phenoxy]furo[3,2-c]pyridine, D(1A) dopamine receptor, Fab 8D3 heavy chain, ... | | Authors: | Zhuang, Y, Xu, Y, Fan, L, Wang, S, Xu, H.E. | | Deposit date: | 2023-07-01 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of psychedelic LSD recognition at dopamine D 1 receptor.
Neuron, 112, 2024
|
|
8JXR
 
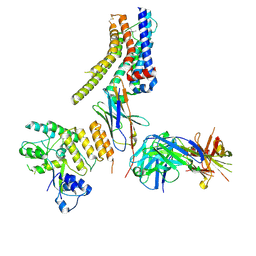 | | Structure of nanobody-bound DRD1_LSD complex | | Descriptor: | (8alpha)-N,N-diethyl-6-methyl-9,10-didehydroergoline-8-carboxamide, D(1A) dopamine receptor, Fab 8D3 heavy chain, ... | | Authors: | Zhuang, Y, Xu, Y, Fan, L, Wang, S, Xu, H.E. | | Deposit date: | 2023-07-01 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structural basis of psychedelic LSD recognition at dopamine D 1 receptor.
Neuron, 112, 2024
|
|
6JHY
 
 | |
8SMV
 
 | | GPR161 Gs heterotrimer | | Descriptor: | CHOLESTEROL, G-protein coupled receptor 161, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hoppe, N, Manglik, A, Harrison, S. | | Deposit date: | 2023-04-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | GPR161 structure uncovers the redundant role of sterol-regulated ciliary cAMP signaling in the Hedgehog pathway.
Nat.Struct.Mol.Biol., 31, 2024
|
|
1C6X
 
 | |
1C6Y
 
 | |
7WZ2
 
 | | SARS-CoV-2 (D614G) Spike trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-02-16 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural Study of SARS-CoV-2 Antibodies Identifies a Broad-Spectrum Antibody That Neutralizes the Omicron Variant by Disassembling the Spike Trimer.
J.Virol., 96, 2022
|
|
7WZ1
 
 | | SARS-CoV-2 Omicron Spike trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-02-16 | | Release date: | 2022-07-27 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Study of SARS-CoV-2 Antibodies Identifies a Broad-Spectrum Antibody That Neutralizes the Omicron Variant by Disassembling the Spike Trimer.
J.Virol., 96, 2022
|
|
7Y0Z
 
 | | Crystal structure of Pseudomonas aeruginosa PvrA | | Descriptor: | TetR family transcriptional regulator | | Authors: | Liang, H, Zhang, Q, Bartlam, M. | | Deposit date: | 2022-06-06 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Regulatory and structural mechanisms of PvrA-mediated regulation of the PQS quorum-sensing system and PHA biosynthesis in Pseudomonas aeruginosa.
Nucleic Acids Res., 51, 2023
|
|
7Y0Y
 
 | | Crystal structure of Pseudomonas aeruginosa PvrA (SeMet) | | Descriptor: | TetR family transcriptional regulator | | Authors: | Liang, H, Zhang, Q, Bartlam, M. | | Deposit date: | 2022-06-06 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Regulatory and structural mechanisms of PvrA-mediated regulation of the PQS quorum-sensing system and PHA biosynthesis in Pseudomonas aeruginosa.
Nucleic Acids Res., 51, 2023
|
|
7WZX
 
 | | The structure of a Twitch Radical SAM Dehydrogenase SpeY | | Descriptor: | (2~{S},4~{S},6~{R})-2-[(2~{S},3~{R},5~{S},6~{R})-3,5-bis(methylamino)-2,4,6-tris(oxidanyl)cyclohexyl]oxy-6-methyl-4-oxidanyl-oxan-3-one, 4Fe-4S cluster-binding domain-containing protein, GLYCEROL, ... | | Authors: | Hou, X.L, Zhou, J.H. | | Deposit date: | 2022-02-19 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.980013 Å) | | Cite: | Dioxane Bridge Formation during the Biosynthesis of Spectinomycin Involves a Twitch Radical S -Adenosyl Methionine Dehydrogenase That May Have Evolved from an Epimerase.
J.Am.Chem.Soc., 144, 2022
|
|
7X0B
 
 | | The structure of a Twitch Radical SAM Dehydrogenase SpeY | | Descriptor: | 4Fe-4S cluster-binding domain-containing protein, CHLORIDE ION, GLYCEROL, ... | | Authors: | Hou, X.L, Zhou, J.H. | | Deposit date: | 2022-02-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.02027535 Å) | | Cite: | Dioxane Bridge Formation during the Biosynthesis of Spectinomycin Involves a Twitch Radical S -Adenosyl Methionine Dehydrogenase That May Have Evolved from an Epimerase.
J.Am.Chem.Soc., 144, 2022
|
|
7WZV
 
 | | The structure of a Twitch Radical SAM Dehydrogenase SpeY | | Descriptor: | (1~{S},2~{R},4~{S},5~{R})-2,4-bis(methylamino)-6-[(2~{S},3~{R},4~{S},6~{R})-6-methyl-3,4-bis(oxidanyl)oxan-2-yl]oxy-cyclohexane-1,3,5-triol, 1,2-ETHANEDIOL, 4Fe-4S cluster-binding domain-containing protein, ... | | Authors: | Zhou, J.H, Hou, X.L. | | Deposit date: | 2022-02-19 | | Release date: | 2022-12-28 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.899313 Å) | | Cite: | Dioxane Bridge Formation during the Biosynthesis of Spectinomycin Involves a Twitch Radical S -Adenosyl Methionine Dehydrogenase That May Have Evolved from an Epimerase.
J.Am.Chem.Soc., 144, 2022
|
|
7YC9
 
 | | Co-crystal structure of BTK kinase domain with inhibitor | | Descriptor: | (7~{S})-2-(4-bromanyl-3,5-dimethoxy-phenyl)-7-(1-propanoylpiperidin-4-yl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide, 1,2-ETHANEDIOL, Tyrosine-protein kinase BTK | | Authors: | Zhou, X. | | Deposit date: | 2022-07-01 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery of BGB-8035, a Highly Selective Covalent Inhibitor of Bruton's Tyrosine Kinase for B-Cell Malignancies and Autoimmune Diseases.
J.Med.Chem., 66, 2023
|
|
6XEY
 
 | | Cryo-EM structure of the SARS-CoV-2 spike glycoprotein bound to Fab 2-4 | | Descriptor: | 2-4 Heavy Chain, 2-4 Light Chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rapp, M, Shapiro, L, Ho, D.D. | | Deposit date: | 2020-06-14 | | Release date: | 2020-07-22 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike.
Nature, 584, 2020
|
|
1DJN
 
 | | STRUCTURAL AND BIOCHEMICAL CHARACTERIZATION OF RECOMBINANT WILD TYPE TRIMETHYLAMINE DEHYDROGENASE FROM METHYLOPHILUS METHYLOTROPHUS (SP. W3A1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Trickey, P, Basran, J, Lian, L.-Y, Chen, Z.-W, Barton, J.D, Sutcliffe, M.J, Scrutton, N.S, Mathews, F.S. | | Deposit date: | 1999-12-03 | | Release date: | 1999-12-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical characterization of recombinant wild type and a C30A mutant of trimethylamine dehydrogenase from methylophilus methylotrophus (sp. W(3)A(1)).
Biochemistry, 39, 2000
|
|
1DJQ
 
 | | STRUCTURAL AND BIOCHEMICAL CHARACTERIZATION OF RECOMBINANT C30A MUTANT OF TRIMETHYLAMINE DEHYDROGENASE FROM METHYLOPHILUS METHYLOTROPHUS (SP. W3A1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Trickey, P, Basran, J, Lian, L.-Y, Chen, Z.-W, Barton, J.D, Sutcliffe, M.J, Scrutton, N.S, Mathews, F.S. | | Deposit date: | 1999-12-03 | | Release date: | 1999-12-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical characterization of recombinant wild type and a C30A mutant of trimethylamine dehydrogenase from methylophilus methylotrophus (sp. W(3)A(1)).
Biochemistry, 39, 2000
|
|
4QP0
 
 | | Crystal Structure Analysis of the Endo-1,4-beta-mannanase from Rhizomucor miehei | | Descriptor: | Endo-beta-mannanase, SULFATE ION | | Authors: | Zheng, Q.J, Peng, Z, Liu, Y, Yan, Q.J, Chen, Z.Z, Qin, Z. | | Deposit date: | 2014-06-22 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7WY5
 
 | | ADGRL3/Gq complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Isoform 3 of Adhesion G protein-coupled receptor L3, ... | | Authors: | He, Y, Qian, Y. | | Deposit date: | 2022-02-15 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural insights into adhesion GPCR ADGRL3 activation and Gq, Gs, Gi, and G12 coupling.
Mol.Cell, 82, 2022
|
|
7WYB
 
 | | ADGRL3/Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | He, Y, Qian, Y. | | Deposit date: | 2022-02-15 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural insights into adhesion GPCR ADGRL3 activation and G q , G s , G i , and G 12 coupling.
Mol.Cell, 82, 2022
|
|
7WY8
 
 | | ADGRL3/Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Isoform 3 of Adhesion G protein-coupled receptor L3, ... | | Authors: | He, Y, Qian, Y. | | Deposit date: | 2022-02-15 | | Release date: | 2022-10-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural insights into adhesion GPCR ADGRL3 activation and Gq, Gs, Gi, and G12 coupling
Mol.Cell, 82, 2022
|
|
7X10
 
 | | ADGRL3/miniG12 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Isoform 3 of Adhesion G protein-coupled receptor L3, ... | | Authors: | He, Y, Qian, Y. | | Deposit date: | 2022-02-22 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural insights into adhesion GPCR ADGRL3 activation and Gq, Gs, Gi, and G12 coupling.
Mol.Cell, 82, 2022
|
|
2MTA
 
 | | CRYSTAL STRUCTURE OF A TERNARY ELECTRON TRANSFER COMPLEX BETWEEN METHYLAMINE DEHYDROGENASE, AMICYANIN AND A C-TYPE CYTOCHROME | | Descriptor: | AMICYANIN, COPPER (II) ION, CYTOCHROME C551I, ... | | Authors: | Chen, L, Mathews, F.S. | | Deposit date: | 1993-10-26 | | Release date: | 1994-01-31 | | Last modified: | 2021-03-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of an electron transfer complex: methylamine dehydrogenase, amicyanin, and cytochrome c551i.
Science, 264, 1994
|
|
5IHC
 
 | | MELK in complex with NVS-MELK12B | | Descriptor: | 4-[1-(2-fluorophenyl)-1H-pyrazol-4-yl]-3-[(piperidin-4-yl)methoxy]pyridine, Maternal embryonic leucine zipper kinase | | Authors: | Sprague, E.R, Brazell, T. | | Deposit date: | 2016-02-29 | | Release date: | 2016-06-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Toward the Validation of Maternal Embryonic Leucine Zipper Kinase: Discovery, Optimization of Highly Potent and Selective Inhibitors, and Preliminary Biology Insight.
J.Med.Chem., 59, 2016
|
|
