8E6U
 
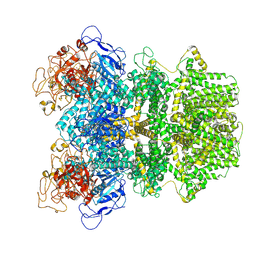 | | Human TRPM2 ion channel in 1 mM F-dADPR | | Descriptor: | Transient receptor potential cation channel subfamily M member 2, [(2R,3S,5R)-5-(6-amino-8-phenyl-9H-purin-9-yl)-3-hydroxyoxolan-2-yl]methyl [(2R,3S,4S,5R)-3,4,5-trihydroxyoxolan-2-yl]methyl dihydrogen diphosphate (non-preferred name) | | Authors: | Wang, L, Fu, T.M, Xia, S, Wu, H. | | Deposit date: | 2022-08-23 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A unified mechanism for human TRPM2 activation, desensitization and inhibition
To Be Published
|
|
8E6Q
 
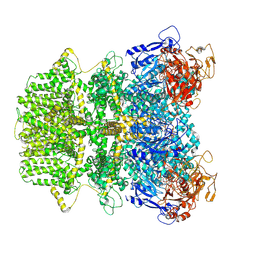 | | Human TRPM2 ion channel in 1 mM ADPR | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Transient receptor potential cation channel subfamily M member 2 | | Authors: | Wang, L, Fu, T.M, Xia, S, Wu, H. | | Deposit date: | 2022-08-23 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A unified mechanism for human TRPM2 activation, desensitization and inhibition
To Be Published
|
|
8E6R
 
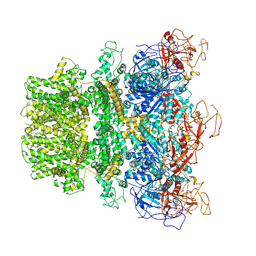 | | Human TRPM2 ion channel in 1 mM dADPR | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Transient receptor potential cation channel subfamily M member 2, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Wang, L, Fu, T.M, Xia, S, Wu, H. | | Deposit date: | 2022-08-23 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | A unified mechanism for human TRPM2 activation, desensitization and inhibition
To Be Published
|
|
8E6T
 
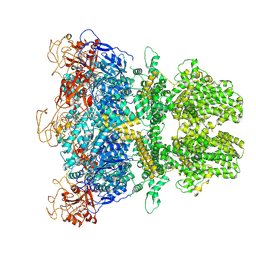 | | Human TRPM2 ion channel in 1 mM BR-ADPR | | Descriptor: | (3R,4R,5R)-5-(6-amino-8-bromo-9H-purin-9-yl)-3,4,5-trihydroxypentyl [(2S,3S,4S,5R)-3,4,5-trihydroxyoxolan-2-yl]methyl dihydrogen diphosphate (non-preferred name), Transient receptor potential cation channel subfamily M member 2 | | Authors: | Wang, L, Fu, T.M, Xia, S, Wu, H. | | Deposit date: | 2022-08-23 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A unified mechanism for human TRPM2 activation, desensitization and inhibition
To Be Published
|
|
8E6S
 
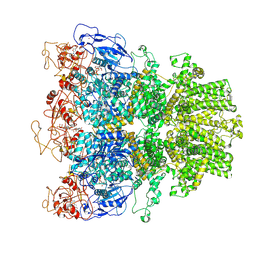 | | Human TRPM2 ion channel in 1 mM dADPR and Ca2+ | | Descriptor: | 2'-DEOXYADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5-DIPHOSPHORIBOSE, CALCIUM ION, ... | | Authors: | Wang, L, Fu, T.M, Xia, S, Wu, H. | | Deposit date: | 2022-08-23 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | A unified mechanism for human TRPM2 activation, desensitization and inhibition
To Be Published
|
|
6IRF
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 6.3, Class I | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | Authors: | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
5TUY
 
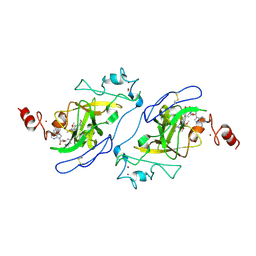 | | Structure of human G9a SET-domain (EHMT2) in complex with inhibitor MS0124 | | Descriptor: | 6,7-dimethoxy-N-(1-methylpiperidin-4-yl)-2-(morpholin-4-yl)quinazolin-4-amine, Histone-lysine N-methyltransferase EHMT2, S-ADENOSYLMETHIONINE, ... | | Authors: | Babault, N, Xiong, Y, Liu, J, Jin, J. | | Deposit date: | 2016-11-07 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of Potent and Selective Inhibitors for G9a-Like Protein (GLP) Lysine Methyltransferase.
J. Med. Chem., 60, 2017
|
|
5VSC
 
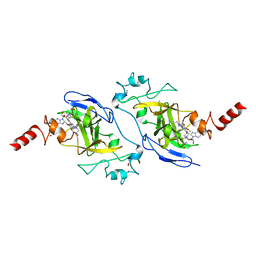 | | Structure of human G9a SET-domain (EHMT2) in complex with inhibitor 13 | | Descriptor: | 6,7-dimethoxy-N~2~-methyl-N~4~-(1-methylpiperidin-4-yl)-N~2~-propylquinazoline-2,4-diamine, Histone-lysine N-methyltransferase EHMT2, S-ADENOSYLMETHIONINE, ... | | Authors: | Babault, N, Xiong, Y, Liu, J, Jin, J. | | Deposit date: | 2017-05-11 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-activity relationship studies of G9a-like protein (GLP) inhibitors.
Bioorg. Med. Chem., 25, 2017
|
|
5VSF
 
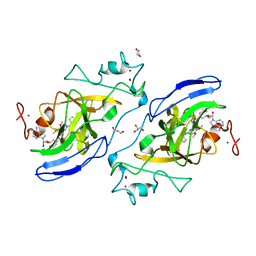 | | Structure of human GLP SET-domain (EHMT1) in complex with inhibitor 17 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, GLYCEROL, Histone-lysine N-methyltransferase EHMT1, ... | | Authors: | Babault, N, Xiong, Y, Liu, J, Jin, J. | | Deposit date: | 2017-05-11 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-activity relationship studies of G9a-like protein (GLP) inhibitors.
Bioorg. Med. Chem., 25, 2017
|
|
4BV6
 
 | | Refined crystal structure of the human Apoptosis inducing factor | | Descriptor: | APOPTOSIS-INDUCING FACTOR 1, MITOCHONDRIAL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Martinez-Julvez, M, Herguedas, B, Hermoso, J.A, Ferreira, P, Villanueva, R, Medina, M. | | Deposit date: | 2013-06-25 | | Release date: | 2014-09-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights Into the Coenzyme Mediated Monomer-Dimer Transition of the Pro-Apoptotic Apoptosis Inducing Factor.
Biochemistry, 53, 2014
|
|
4JHC
 
 | | Crystal structure of the uncharacterized Maf protein YceF from E. coli | | Descriptor: | GLYCEROL, Maf-like protein YceF, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Xu, X, Cui, H, Tchigvintsev, A, Flick, R, Brown, G, Popovic, A, Yakunin, A.F, Savchenko, A. | | Deposit date: | 2013-03-04 | | Release date: | 2013-03-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biochemical and structural studies of conserved maf proteins revealed nucleotide pyrophosphatases with a preference for modified nucleotides.
Chem.Biol., 20, 2013
|
|
3SMQ
 
 | | Crystal structure of protein arginine methyltransferase 3 | | Descriptor: | 1-(1,2,3-benzothiadiazol-6-yl)-3-[2-(cyclohex-1-en-1-yl)ethyl]urea, CHLORIDE ION, Protein arginine N-methyltransferase 3, ... | | Authors: | Dobrovetsky, E, Dong, A, Walker, J.R, Siarheyeva, A, Senisterra, G, Wasney, G.A, Smil, D, Bolshan, Y, Nguyen, K.T, Allali-Hassani, A, Hajian, T, Poda, G, Bountra, C, Weigelt, J, Edwards, A.M, Al-Awar, R, Brown, P.J, Schapira, M, Arrowsmith, C.H, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-06-28 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An allosteric inhibitor of protein arginine methyltransferase 3.
Structure, 20, 2012
|
|
2MEZ
 
 | | Flexible anchoring of archaeal MBF1 on ribosomes suggests role as recruitment factor | | Descriptor: | Multiprotein Bridging Factor (MBP-like) | | Authors: | Launay, H, Blombarch, F, Camilloni, C, Vendruscolo, M, van des Oost, J, Christodoulou, J. | | Deposit date: | 2013-10-03 | | Release date: | 2014-06-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Archaeal MBF1 binds to 30S and 70S ribosomes via its helix-turn-helix domain.
Biochem.J., 462, 2014
|
|
9FEB
 
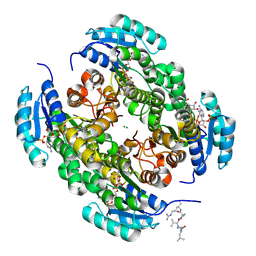 | |
5IN1
 
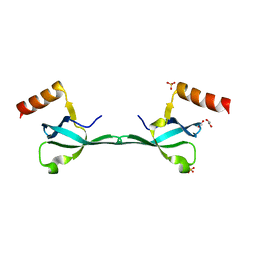 | | Crystal Structure of the MRG701 chromodomain | | Descriptor: | 1,2-ETHANEDIOL, MRG701, SULFATE ION | | Authors: | Huang, Y, Liu, Y. | | Deposit date: | 2016-03-07 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural studies on MRG701 chromodomain reveal a novel dimerization interface of MRG proteins in green plants
Protein Cell, 7, 2016
|
|
9FE6
 
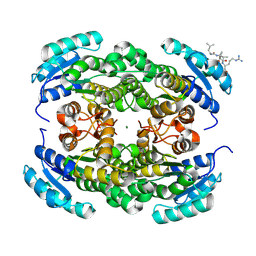 | | Short-chain dehydrogenase/reductase (SDR) from Thermus caliditerrae | | Descriptor: | MAGNESIUM ION, SDR family oxidoreductase | | Authors: | Kapur, B, Nar, H. | | Deposit date: | 2024-05-17 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.875 Å) | | Cite: | In silico enzyme screening identifies an SDR ketoreductase from Thermus caliditerrae as an attractive biocatalyst and promising candidate for protein engineering
Front Chem Biol, 2024
|
|
7F9O
 
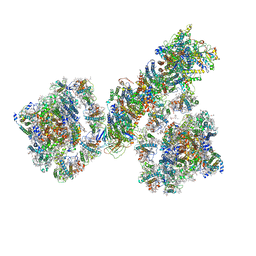 | | PSI-NDH supercomplex of Barley | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Wang, W.D, Shen, L, Tang, K, Han, G.Y, Shen, J.R, Zhang, X. | | Deposit date: | 2021-07-04 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Architecture of the chloroplast PSI-NDH supercomplex in Hordeum vulgare.
Nature, 601, 2022
|
|
7L0O
 
 | |
5J87
 
 | |
5KCV
 
 | | Crystal structure of allosteric inhibitor, ARQ 092, in complex with autoinhibited form of AKT1 | | Descriptor: | 3-[3-[4-(1-azanylcyclobutyl)phenyl]-5-phenyl-imidazo[4,5-b]pyridin-2-yl]pyridin-2-amine, RAC-alpha serine/threonine-protein kinase | | Authors: | Eathiraj, S. | | Deposit date: | 2016-06-07 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of 3-(3-(4-(1-Aminocyclobutyl)phenyl)-5-phenyl-3H-imidazo[4,5-b]pyridin-2-yl)pyridin-2-amine (ARQ 092): An Orally Bioavailable, Selective, and Potent Allosteric AKT Inhibitor.
J.Med.Chem., 59, 2016
|
|
9CJL
 
 | | Molecular basis of TMED9 dodecamer | | Descriptor: | Transmembrane emp24 domain-containing protein 9, [(2R)-2-[(E)-octadec-9-enoyl]oxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] (E)-octadec-9-enoate | | Authors: | Le, X, Xiong, P. | | Deposit date: | 2024-07-06 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Molecular basis of TMED9 oligomerization and entrapment of misfolded protein cargo in the early secretory pathway.
Sci Adv, 10, 2024
|
|
9CJK
 
 | | Human TMED9 octamer structure | | Descriptor: | Transmembrane emp24 domain-containing protein 9, [(2R)-2-[(E)-octadec-9-enoyl]oxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] (E)-octadec-9-enoate | | Authors: | Le, X, Xiong, P. | | Deposit date: | 2024-07-06 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular basis of TMED9 oligomerization and entrapment of misfolded protein cargo in the early secretory pathway.
Sci Adv, 10, 2024
|
|
3KXL
 
 | | crystal structure of SsGBP mutation variant G235S | | Descriptor: | GTP-binding protein (HflX), THIOCYANATE ION | | Authors: | Huang, B, Li, X, Zhang, X.C, Rao, Z. | | Deposit date: | 2009-12-03 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Functional study on GTP hydrolysis by the GTP binding protein from Sulfolobus solfataricus, a member of the HflX family.
J.Biochem., 2010
|
|
3KXI
 
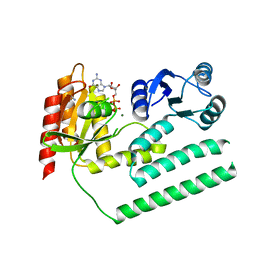 | | crystal structure of SsGBP and GDP complex | | Descriptor: | GTP-binding protein (HflX), GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, B, Li, X, Zhang, X.C, Rao, Z. | | Deposit date: | 2009-12-03 | | Release date: | 2010-05-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Functional study on GTP hydrolysis by the GTP binding protein from Sulfolobus solfataricus, a member of the HflX family.
J.Biochem., 2010
|
|
3I6G
 
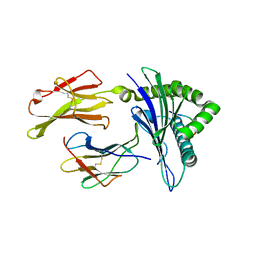 | | Newly identified epitope Mn2 from SARS-CoV M protein complexed withHLA-A*0201 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Liu, J. | | Deposit date: | 2009-07-07 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | The membrane protein of severe acute respiratory syndrome coronavirus acts as a dominant immunogen revealed by a clustering region of novel functionally and structurally defined cytotoxic T-lymphocyte epitopes.
J.INFECT.DIS., 202, 2010
|
|
