4ZI7
 
 | | CRYSTAL STRUCTURE OF TUBULIN-STATHMIN-TTL-HTI286 COMPLEX | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Wang, Y, Zhang, R. | | Deposit date: | 2015-04-27 | | Release date: | 2016-07-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural Insights into the Pharmacophore of Vinca Domain Inhibitors of Microtubules
Mol.Pharmacol., 89, 2016
|
|
4RAY
 
 | | Crystal structure of Magnetospirillum gryphiswaldense MSR-1 Apo-Fur | | Descriptor: | CITRATE ANION, DNA-binding transcriptional dual regulator of siderophore biosynthesis and transport(Fur family), SULFATE ION | | Authors: | Deng, Z, Liu, Z, Chen, Z. | | Deposit date: | 2014-09-12 | | Release date: | 2015-07-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Mechanistic insights into metal ion activation and operator recognition by the ferric uptake regulator.
Nat Commun, 6
|
|
9JM0
 
 | | retron Ec86-effector fiber | | Descriptor: | DNA (85-MER), NICOTINAMIDE, RNA (5'-R(P*CP*GP*UP*AP*AP*GP*GP*GP*UP*GP*CP*GP*CP*A)-3'), ... | | Authors: | Wang, Y.J, Guan, Z.Y, Wang, C, Zou, T.T. | | Deposit date: | 2024-09-20 | | Release date: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | DNA methylation activates retron Ec86 filaments for antiphage defense.
Cell Rep, 43, 2024
|
|
4RB3
 
 | |
4RB0
 
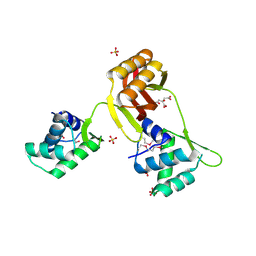 | | Crystal structure of Magnetospirillum gryphiswaldense MSR-1 SeMet-Apo-Fur | | Descriptor: | CITRATE ANION, DNA-binding transcriptional dual regulator of siderophore biosynthesis and transport(Fur family), SULFATE ION | | Authors: | Deng, Z, Chen, Z. | | Deposit date: | 2014-09-12 | | Release date: | 2015-07-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanistic insights into metal ion activation and operator recognition by the ferric uptake regulator.
Nat Commun, 6
|
|
4RB1
 
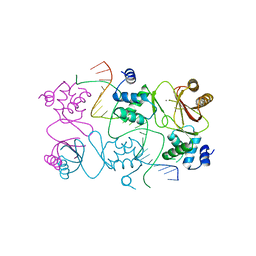 | | Crystal structure of Magnetospirillum gryphiswaldense MSR-1 Fur-Mn2+-E. coli Fur box | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*TP*AP*AP*TP*GP*AP*TP*AP*AP*TP*CP*AP*TP*TP*AP*TP*CP*CP*GP*C)-3'), DNA-binding transcriptional dual regulator of siderophore biosynthesis and transport(Fur family), MANGANESE (II) ION | | Authors: | Deng, Z, Chen, Z. | | Deposit date: | 2014-09-12 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Mechanistic insights into metal ion activation and operator recognition by the ferric uptake regulator.
Nat Commun, 6
|
|
4TVZ
 
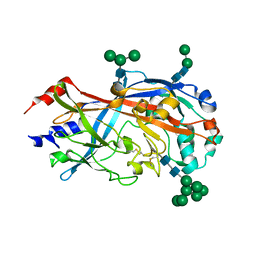 | | Crystal Structure of SCARB2 in Neural Condition (pH7.5) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Scavenger receptor class B member 2, ... | | Authors: | Dang, M.H, Wang, X.X, Rao, Z.H. | | Deposit date: | 2014-06-29 | | Release date: | 2015-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.006 Å) | | Cite: | Molecular mechanism of SCARB2-mediated attachment and uncoating of EV71
Protein Cell, 5, 2014
|
|
4TW0
 
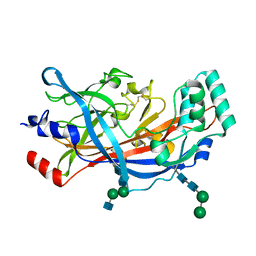 | | Crystal Structure of SCARB2 in Acidic Condition (pH4.8) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dang, M.H, Wang, X.X, Rao, Z.H. | | Deposit date: | 2014-06-29 | | Release date: | 2015-07-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.648 Å) | | Cite: | Molecular mechanism of SCARB2-mediated attachment and uncoating of EV71
Protein Cell, 5, 2014
|
|
6M79
 
 | |
6J6M
 
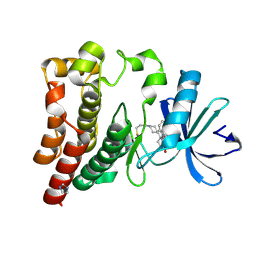 | | Co-crystal structure of BTK kinase domain with Zanubrutinib | | Descriptor: | (7S)-2-(4-phenoxyphenyl)-7-(1-propanoylpiperidin-4-yl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide, IMIDAZOLE, Tyrosine-protein kinase BTK | | Authors: | Zhou, X, Hong, Y. | | Deposit date: | 2019-01-15 | | Release date: | 2019-10-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Discovery of Zanubrutinib (BGB-3111), a Novel, Potent, and Selective Covalent Inhibitor of Bruton's Tyrosine Kinase.
J.Med.Chem., 62, 2019
|
|
6LN1
 
 | | A natural inhibitor of DYRK1A for treatment of diabetes mellitus | | Descriptor: | 1,3,5,8-tetrakis(oxidanyl)xanthen-9-one, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Li, H, Chen, L.X, Zheng, M.Z, Zhang, Q.Z, Zhang, C.L, Wu, C.R, Yang, K.Y, Song, Z.R, Wang, Q.Q, Li, C, Zhou, Y.R, Chen, J.C. | | Deposit date: | 2019-12-28 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | A natural DYRK1A inhibitor as a potential stimulator for beta-cell proliferation in diabetes.
Clin Transl Med, 11, 2021
|
|
8GT6
 
 | | human STING With agonist HB3089 | | Descriptor: | 1-[(2E)-4-{5-carbamoyl-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-[3-(morpholin-4-yl)propoxy]-1H-benzimidazol-1-yl}but-2-en-1-yl]-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-methyl-1H-furo[3,2-e]benzimidazole-5-carboxamide, Stimulator of interferon genes protein | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2022-09-07 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural insights into a shared mechanism of human STING activation by a potent agonist and an autoimmune disease-associated mutation.
Cell Discov, 8, 2022
|
|
8GSZ
 
 | | Structure of STING SAVI-related mutant V147L | | Descriptor: | Stimulator of interferon genes protein | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2022-09-07 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Structural insights into a shared mechanism of human STING activation by a potent agonist and an autoimmune disease-associated mutation.
Cell Discov, 8, 2022
|
|
9II5
 
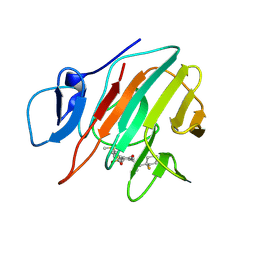 | | Crystal structure of human TRIM21 PRYSPRY in complex with compound 1 | | Descriptor: | E3 ubiquitin-protein ligase TRIM21, ~{N}-[(1-fluoranylcyclohexyl)methyl]-~{N}-methyl-4-(2-methylsulfanylphenyl)-2-methylsulfonyl-benzamide | | Authors: | Zhang, L.Y. | | Deposit date: | 2024-06-19 | | Release date: | 2025-05-07 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Chemically Induced Nuclear Pore Complex Protein Degradation via TRIM21.
Acs Chem.Biol., 20, 2025
|
|
4Y7R
 
 | | Crystal structure of WDR5 in complex with MYC MbIIIb peptide | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MYC MbIIIb peptide, ... | | Authors: | Sun, Q, Phan, J, Olejniczak, E.T, Thomas, L.R, Fesik, S.W, Tansey, W.P. | | Deposit date: | 2015-02-16 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Interaction with WDR5 Promotes Target Gene Recognition and Tumorigenesis by MYC.
Mol.Cell, 58, 2015
|
|
5OVQ
 
 | |
6LNA
 
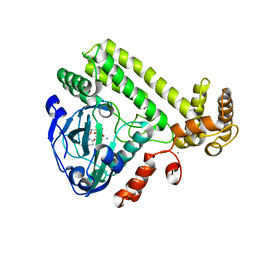 | | YdiU complex with AMPNPP and Mn2+ | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, B, Yang, Y, Ma, Y. | | Deposit date: | 2019-12-28 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | The YdiU Domain Modulates Bacterial Stress Signaling through Mn 2+ -Dependent UMPylation.
Cell Rep, 32, 2020
|
|
5X4S
 
 | | Structure of the N-terminal domain (NTD)of SARS-CoV spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yuan, Y, Zhang, Y, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2017-02-14 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5XGR
 
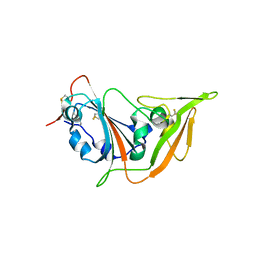 | | Structure of the S1 subunit C-terminal domain from bat-derived coronavirus HKU5 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1 | | Authors: | Xue, H, Qi, J, Song, H, Qihui, W, Shi, Y, Gao, G.F. | | Deposit date: | 2017-04-16 | | Release date: | 2017-05-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the S1 subunit C-terminal domain from bat-derived coronavirus HKU5 spike protein
Virology, 507, 2017
|
|
6KXW
 
 | | Crystal structure of human aquaporin AQP7 in bound to glycerol | | Descriptor: | Aquaporin-7, GLYCEROL | | Authors: | Zhang, L, Yao, D, Zhou, F, Zhang, Q, Zhou, L, Cao, Y. | | Deposit date: | 2019-09-13 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | The structural basis for glycerol permeation by human AQP7
Sci Bull (Beijing), 66, 2020
|
|
6L8Q
 
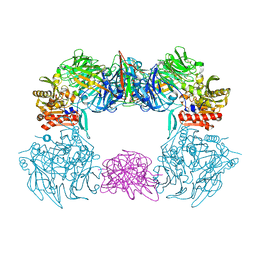 | | Complex structure of bat CD26 and MERS-RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | Authors: | Yuan, Y. | | Deposit date: | 2019-11-07 | | Release date: | 2019-12-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular Basis of Binding between Middle East Respiratory Syndrome Coronavirus and CD26 from Seven Bat Species.
J.Virol., 94, 2020
|
|
6R7Z
 
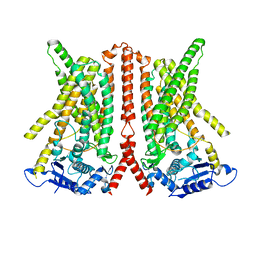 | | CryoEM structure of calcium-free human TMEM16K / Anoctamin 10 in detergent (closed form) | | Descriptor: | Anoctamin-10 | | Authors: | Pike, A.C.W, Bushell, S.R, Shintre, C.A, Tessitore, A, Chu, A, Mukhopadhyay, S, Shrestha, L, Chalk, R, Burgess-Brown, N.A, Love, J, Huiskonen, J.T, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-29 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (5.14 Å) | | Cite: | The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K.
Nat Commun, 10, 2019
|
|
3LFM
 
 | |
7WED
 
 | |
7WEA
 
 | |
