7W0L
 
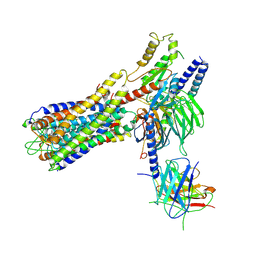 | | Cryo-EM structure of a dimeric GPCR-Gi complex with small molecule | | Descriptor: | (1R,2S)-N-[4-(2,6-dimethoxyphenyl)-5-(6-methylpyridin-2-yl)-1,2,4-triazol-3-yl]-1-(5-methylpyrimidin-2-yl)-1-oxidanyl-propane-2-sulfonamide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yue, Y, Liu, L.E, Wu, L.J, Xu, F, Hanson, M. | | Deposit date: | 2021-11-18 | | Release date: | 2022-07-27 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structural insight into apelin receptor-G protein stoichiometry.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7W0O
 
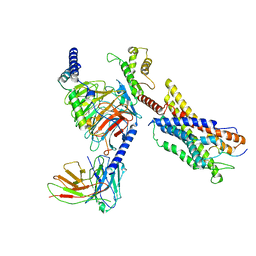 | | Cryo-EM structure of a monomeric GPCR-Gi complex with peptide | | Descriptor: | Apelin receptor early endogenous ligand, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, F, Yue, Y, Liu, L.E, Wu, L.J, Hanson, M. | | Deposit date: | 2021-11-18 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Structural insight into apelin receptor-G protein stoichiometry.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7W0M
 
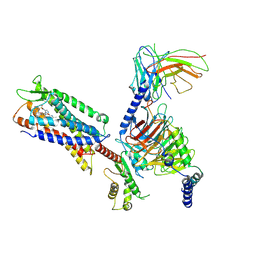 | | Cryo-EM structure of a monomeric GPCR-Gi complex with small molecule | | Descriptor: | (1R,2S)-N-[4-(2,6-dimethoxyphenyl)-5-(6-methylpyridin-2-yl)-1,2,4-triazol-3-yl]-1-(5-methylpyrimidin-2-yl)-1-oxidanyl-propane-2-sulfonamide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, F, Yue, Y, Liu, L.E, Wu, L.J, Hanson, M. | | Deposit date: | 2021-11-18 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural insight into apelin receptor-G protein stoichiometry.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7W0P
 
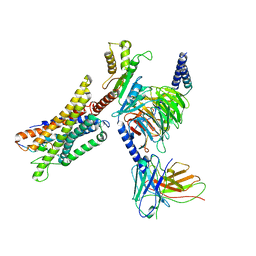 | | Cryo-EM structure of a GPCR-Gi complex with peptide | | Descriptor: | Apelin receptor early endogenous ligand, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, F, Yue, Y, Liu, L.E, Wu, L.J, Hanson, M. | | Deposit date: | 2021-11-18 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural insight into apelin receptor-G protein stoichiometry.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7EVR
 
 | | Crystal structure of hnRNP L RRM2 in complex with SETD2 | | Descriptor: | Heterogeneous nuclear ribonucleoprotein L, SHI domain from Histone-lysine N-methyltransferase SETD2 | | Authors: | Li, F.D, Wang, S.M. | | Deposit date: | 2021-05-22 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the interaction between SETD2 methyltransferase and hnRNP L paralogs for governing co-transcriptional splicing.
Nat Commun, 12, 2021
|
|
7EVS
 
 | | Crystal structure of hnRNP LL RRM2 in complex with SETD2 | | Descriptor: | Heterogeneous nuclear ribonucleoprotein L-like, SHI domain from Histone-lysine N-methyltransferase SETD2, SULFATE ION | | Authors: | Li, F.D, Wang, S.M. | | Deposit date: | 2021-05-22 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of the interaction between SETD2 methyltransferase and hnRNP L paralogs for governing co-transcriptional splicing.
Nat Commun, 12, 2021
|
|
4L0K
 
 | |
5U4X
 
 | | Coactivator-associated arginine methyltransferase 1 with TP-064 | | Descriptor: | Histone-arginine methyltransferase CARM1, N-methyl-N-[(2-{1-[2-(methylamino)ethyl]piperidin-4-yl}pyridin-4-yl)methyl]-3-phenoxybenzamide, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | DONG, A, ZENG, H, Saikatendu, K.S, STONE, H, WALKER, J.R, Seitova, A, Hutchinson, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-12-06 | | Release date: | 2016-12-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | TP-064, a potent and selective small molecule inhibitor of PRMT4 for multiple myeloma.
Oncotarget, 9, 2018
|
|
3SMR
 
 | | Crystal structure of human WD repeat domain 5 with compound | | Descriptor: | 1,2-ETHANEDIOL, 2-chloro-N-[2-(4-methylpiperazin-1-yl)-5-nitrophenyl]benzamide, UNKNOWN ATOM OR ION, ... | | Authors: | Dong, A, Dombrovski, L, Wasney, G.A, Tempel, W, Senisterra, G, Bolshan, Y, Smil, D, Nguyen, K.T, Hajian, T, Poda, G, Al-Awar, R, Bountra, C, Weigelt, J, Edwards, A.M, Brown, P.J, Schapira, M, Arrowsmith, C.H, Vedadi, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-06-28 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Small-molecule inhibition of MLL activity by disruption of its interaction with WDR5.
Biochem. J., 449, 2013
|
|
5TTG
 
 | | Crystal structure of catalytic domain of GLP with MS012 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | DONG, A, ZENG, H, LIU, J, XIONG, Y, BABAULT, N, JIN, J, TEMPEL, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, WU, H, BROWN, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-11-03 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Discovery of Potent and Selective Inhibitors for G9a-Like Protein (GLP) Lysine Methyltransferase.
J. Med. Chem., 60, 2017
|
|
5TEG
 
 | |
5TUY
 
 | | Structure of human G9a SET-domain (EHMT2) in complex with inhibitor MS0124 | | Descriptor: | 6,7-dimethoxy-N-(1-methylpiperidin-4-yl)-2-(morpholin-4-yl)quinazolin-4-amine, Histone-lysine N-methyltransferase EHMT2, S-ADENOSYLMETHIONINE, ... | | Authors: | Babault, N, Xiong, Y, Liu, J, Jin, J. | | Deposit date: | 2016-11-07 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of Potent and Selective Inhibitors for G9a-Like Protein (GLP) Lysine Methyltransferase.
J. Med. Chem., 60, 2017
|
|
5TUZ
 
 | | Structure of human GLP SET-domain (EHMT1) in complex with inhibitor MS0124 | | Descriptor: | 1,2-ETHANEDIOL, 6,7-dimethoxy-N-(1-methylpiperidin-4-yl)-2-(morpholin-4-yl)quinazolin-4-amine, Histone-lysine N-methyltransferase EHMT1, ... | | Authors: | Babault, N, Xiong, Y, Liu, J, Jin, J. | | Deposit date: | 2016-11-07 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of Potent and Selective Inhibitors for G9a-Like Protein (GLP) Lysine Methyltransferase.
J. Med. Chem., 60, 2017
|
|
5VSC
 
 | | Structure of human G9a SET-domain (EHMT2) in complex with inhibitor 13 | | Descriptor: | 6,7-dimethoxy-N~2~-methyl-N~4~-(1-methylpiperidin-4-yl)-N~2~-propylquinazoline-2,4-diamine, Histone-lysine N-methyltransferase EHMT2, S-ADENOSYLMETHIONINE, ... | | Authors: | Babault, N, Xiong, Y, Liu, J, Jin, J. | | Deposit date: | 2017-05-11 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-activity relationship studies of G9a-like protein (GLP) inhibitors.
Bioorg. Med. Chem., 25, 2017
|
|
5VSF
 
 | | Structure of human GLP SET-domain (EHMT1) in complex with inhibitor 17 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, GLYCEROL, Histone-lysine N-methyltransferase EHMT1, ... | | Authors: | Babault, N, Xiong, Y, Liu, J, Jin, J. | | Deposit date: | 2017-05-11 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-activity relationship studies of G9a-like protein (GLP) inhibitors.
Bioorg. Med. Chem., 25, 2017
|
|
5VSE
 
 | | Structure of human G9a SET-domain (EHMT2) in complex with inhibitor 17: N~2~-cyclopentyl-6,7-dimethoxy-N~2~-methyl-N~4~-(1-methylpiperidin-4-yl)quinazoline-2,4-diamine | | Descriptor: | Histone-lysine N-methyltransferase EHMT2, N~2~-cyclopentyl-6,7-dimethoxy-N~2~-methyl-N~4~-(1-methylpiperidin-4-yl)quinazoline-2,4-diamine, S-ADENOSYLMETHIONINE, ... | | Authors: | Babault, N, Xiong, Y, Liu, J, Jin, J. | | Deposit date: | 2017-05-11 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-activity relationship studies of G9a-like protein (GLP) inhibitors.
Bioorg. Med. Chem., 25, 2017
|
|
5VSD
 
 | | Structure of human GLP SET-domain (EHMT1) in complex with inhibitor 13 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 6,7-dimethoxy-N~2~-methyl-N~4~-(1-methylpiperidin-4-yl)-N~2~-propylquinazoline-2,4-diamine, GLYCEROL, ... | | Authors: | Babault, N, Xiong, Y, Liu, J, Jin, J. | | Deposit date: | 2017-05-11 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-activity relationship studies of G9a-like protein (GLP) inhibitors.
Bioorg. Med. Chem., 25, 2017
|
|
3UR4
 
 | | Crystal structure of human WD repeat domain 5 with compound | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Dong, A, Dombrovski, L, Senisterra, G, Wernimont, A, Wasney, G.A, Allali Hassani, A, Nguyen, K.T, Smil, D, Bolshan, Y, Hajian, T, Poda, G, Chau, I, Al-Awar, R, Bountra, C, Weigelt, J, Edwards, A.M, Arrowsmith, C.H, Brown, P, Schapira, M, Vedadi, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-11-21 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Small-molecule inhibition of MLL activity by disruption of its interaction with WDR5.
Biochem. J., 449, 2013
|
|
7ENQ
 
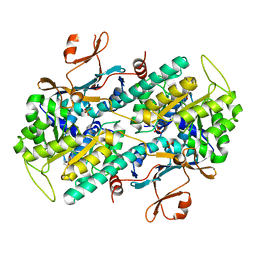 | | Crystal structure of human NAMPT in complex with compound NAT | | Descriptor: | 2-(2-~{tert}-butylphenoxy)-~{N}-(4-hydroxyphenyl)ethanamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Wang, G, Wu, C, Liu, M, Yao, H, Li, C, Wang, L, Tang, Y. | | Deposit date: | 2021-04-19 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.204966 Å) | | Cite: | Discovery of small-molecule activators of nicotinamide phosphoribosyltransferase (NAMPT) and their preclinical neuroprotective activity.
Cell Res., 32, 2022
|
|
8WRZ
 
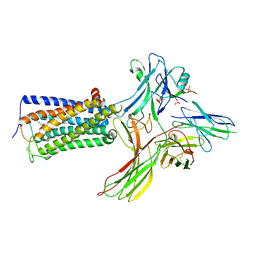 | | Cry-EM structure of cannabinoid receptor-arrestin 2 complex | | Descriptor: | (6~{a}~{R},9~{R},10~{a}~{R})-9-(hydroxymethyl)-3-(8-isothiocyanato-2-methyl-octan-2-yl)-6,6-dimethyl-6~{a},7,8,9,10,10~{a}-hexahydrobenzo[c]chromen-1-ol, Beta-arrestin-1, Soluble cytochrome b562,Cannabinoid receptor 1, ... | | Authors: | Wang, Y.X, Wang, T, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2023-10-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of cannabinoid receptor CB1-beta-arrestin complex.
Protein Cell, 15, 2024
|
|
5Y5P
 
 | |
5Y5Q
 
 | |
5YJ8
 
 | |
5Y5O
 
 | |
5Y0U
 
 | | The solution structure of AEBP2 C2H2 zinc fingers | | Descriptor: | ZINC ION, Zinc finger protein AEBP2 | | Authors: | Sun, A, Shi, Y, Wu, J. | | Deposit date: | 2017-07-18 | | Release date: | 2018-08-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and biochemical insights into human zinc finger protein AEBP2 reveals interactions with RBBP4.
Protein Cell, 9, 2018
|
|
