6LHA
 
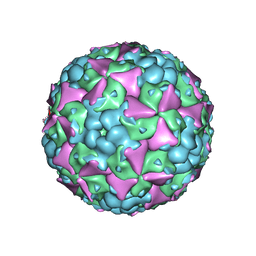 | | The cryo-EM structure of coxsackievirus A16 mature virion | | Descriptor: | SPHINGOSINE, VP1 protein, VP2 protein, ... | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-07 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHQ
 
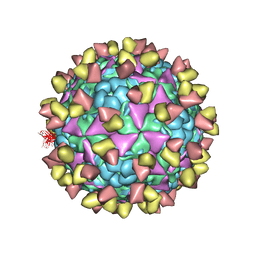 | | The cryo-EM structure of coxsackievirus A16 mature virion in complex with Fab NA9D7 | | Descriptor: | SPHINGOSINE, VP1 protein, VP2 protein, ... | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-09 | | Release date: | 2020-02-05 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHT
 
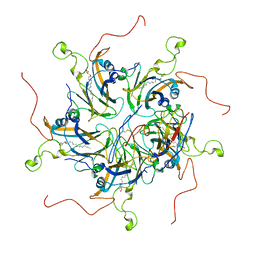 | | Localized reconstruction of coxsackievirus A16 mature virion in complex with Fab 18A7 | | Descriptor: | SPHINGOSINE, VP1 protein, heavy chain variable region of Fab 18A7, ... | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-10 | | Release date: | 2020-02-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
8DXS
 
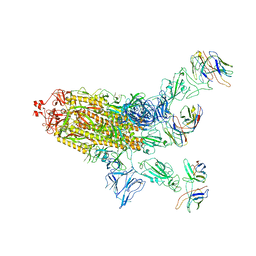 | |
8DW9
 
 | |
8DWA
 
 | |
5JDU
 
 | | Crystal structure for human thrombin mutant D189A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pozzi, N, Chen, Z, Di Cera, E. | | Deposit date: | 2016-04-17 | | Release date: | 2016-07-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Loop Electrostatics Asymmetry Modulates the Preexisting Conformational Equilibrium in Thrombin.
Biochemistry, 55, 2016
|
|
3ZP0
 
 | | INFLUENZA VIRUS (VN1194) H5 HA with LSTa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose | | Authors: | Liu, J, Stevens, D.J, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2013-02-26 | | Release date: | 2013-10-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Changes in the Hemagglutinin of H5N1 Viruses During Human Infection - Influence on Receptor Binding.
Virology, 447, 2013
|
|
3ZWM
 
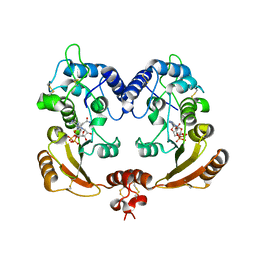 | | Crystal structure of ADP ribosyl cyclase complexed with substrate NAD and product cADPR | | Descriptor: | ADP-RIBOSYL CYLCASE, CYCLIC ADENOSINE DIPHOSPHATE-RIBOSE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kotaka, M, Graeff, R, Zhang, L.H, Lee, H.C, Hao, Q. | | Deposit date: | 2011-08-02 | | Release date: | 2011-11-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Studies of Intermediates Along the Cyclization Pathway of Aplysia Adp-Ribosyl Cyclase.
J.Mol.Biol., 415, 2012
|
|
3ZWO
 
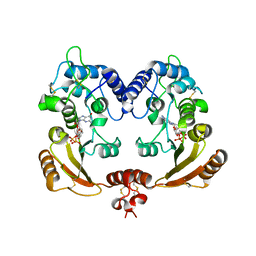 | | Crystal structure of ADP ribosyl cyclase complexed with reaction intermediate | | Descriptor: | 3-(AMINOCARBONYL)-1-[(2R,3R,4S,5R)-5-({[(S)-{[(S)-{[(2R,3S,4R,5R)-5-(2-AMINO-6-OXO-1,6-DIHYDRO-9H-PURIN-9-YL)-3,4-DIHYD ROXYTETRAHYDROFURAN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]OXY}METHYL)-3,4-DIHYDROXYTETRAHYDROFURAN-2- YL]PYRIDINIUM, ADP-RIBOSYL CYCLASE, GUANOSINE DIPHOSPHATE RIBOSE | | Authors: | Kotaka, M, Graeff, R, Zhang, L.H, Lee, H.C, Hao, Q. | | Deposit date: | 2011-08-02 | | Release date: | 2011-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Studies of Intermediates Along the Cyclization Pathway of Aplysia Adp-Ribosyl Cyclase.
J.Mol.Biol., 415, 2012
|
|
3ZWY
 
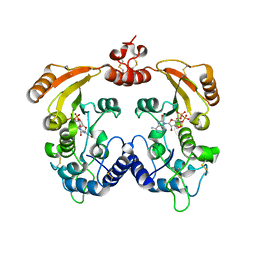 | | Crystal structure of ADP-ribosyl cyclase complexed with 8-bromo-ADP- ribose and cyclic 8-bromo-cyclic-ADP-ribose | | Descriptor: | (2R,3R,4S,5R,13R,14S,15R,16R)-24-amino-18-bromo-3,4,14,15-tetrahydroxy-7,9,11,25,26-pentaoxa-17,19,22-triaza-1-azonia-8 ,10-diphosphapentacyclo[18.3.1.1^2,5^.1^13,16^.0^17,21^]hexacosa-1(24),18,20,22-tetraene-8,10-diolate 8,10-dioxide, ADP-RIBOSYL CYCLASE, [(2R,3S,4R,5R)-5-(6-amino-8-bromo-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3S,4S)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate | | Authors: | Kotaka, M, Graeff, R, Zhang, L.H, Lee, H.C, Hao, Q. | | Deposit date: | 2011-08-03 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Studies of Intermediates Along the Cyclization Pathway of Aplysia Adp-Ribosyl Cyclase.
To be Published
|
|
4LYP
 
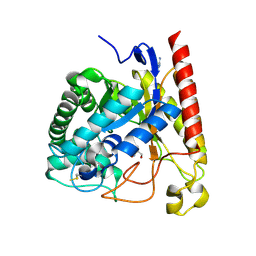 | | Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase from Rhizomucor miehei | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Exo-beta-1,4-mannosidase, GUANIDINE | | Authors: | Jiang, Z.Q, Zhou, P, Yang, S.Q, Liu, Y, Yan, Q.J. | | Deposit date: | 2013-07-31 | | Release date: | 2014-08-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 beta-mannosidase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3ZWX
 
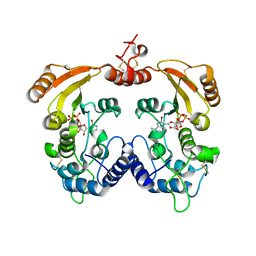 | | Crystal structure of ADP-ribosyl cyclase complexed with 8-bromo-ADP- ribose | | Descriptor: | ADP-RIBOSYL CYCLASE, CHLORIDE ION, [(2R,3S,4R,5R)-5-(6-amino-8-bromo-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3S,4S)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate | | Authors: | Kotaka, M, Graeff, R, Zhang, L.H, Lee, H.C, Hao, Q. | | Deposit date: | 2011-08-03 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Studies of Intermediates Along the Cyclization Pathway of Aplysia Adp-Ribosyl Cyclase.
To be Published
|
|
3ZWW
 
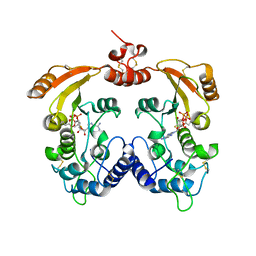 | | Crystal structure of ADP-ribosyl cyclase complexed with ara-2'F-ADP- ribose at 2.3 angstrom | | Descriptor: | ADP-RIBOSYL CYCLASE, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3R,4R)-4-fluoro-3-hydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate | | Authors: | Kotaka, M, Graeff, R, Zhang, L.H, Lee, H.C, Hao, Q. | | Deposit date: | 2011-08-03 | | Release date: | 2011-11-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Studies of Intermediates Along the Cyclization Pathway of Aplysia Adp-Ribosyl Cyclase.
J.Mol.Biol., 415, 2012
|
|
3ZWN
 
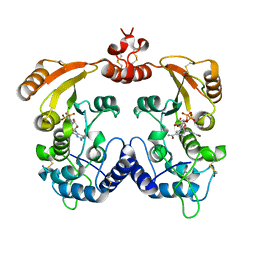 | | Crystal structure of Aplysia cyclase complexed with substrate NGD and product cGDPR | | Descriptor: | 3-(AMINOCARBONYL)-1-[(2R,3R,4S,5R)-5-({[(S)-{[(S)-{[(2R,3S,4R,5R)-5-(2-AMINO-6-OXO-1,6-DIHYDRO-9H-PURIN-9-YL)-3,4-DIHYD ROXYTETRAHYDROFURAN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]OXY}METHYL)-3,4-DIHYDROXYTETRAHYDROFURAN-2- YL]PYRIDINIUM, ADP-RIBOSYL CYCLASE, CYCLIC GUANOSINE DIPHOSPHATE-RIBOSE | | Authors: | Kotaka, M, Graeff, R, Zhang, L.H, Lee, H.C, Hao, Q. | | Deposit date: | 2011-08-02 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Studies of Intermediates Along the Cyclization Pathway of Aplysia Adp-Ribosyl Cyclase.
J.Mol.Biol., 415, 2012
|
|
3ZWV
 
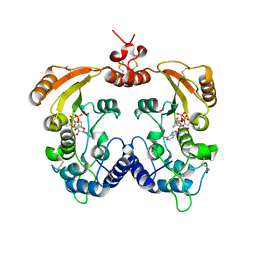 | | Crystal structure of ADP-ribosyl cyclase complexed with ara-2'F-ADP- ribose at 2.3 angstrom | | Descriptor: | ADP-RIBOSYL CYCLASE, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3R,4R)-4-fluoro-3-hydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate | | Authors: | Kotaka, M, Graeff, R, Zhang, L.H, Lee, H.C, Hao, Q. | | Deposit date: | 2011-08-03 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural studies of intermediates along the cyclization pathway of Aplysia ADP-ribosyl cyclase.
J. Mol. Biol., 415, 2012
|
|
3ZWP
 
 | | Crystal structure of ADP ribosyl cyclase complexed with ara-2'F-ADP- ribose at 2.1 angstrom | | Descriptor: | ADP-RIBOSYL CYCLASE, GLYCEROL, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3R,4R)-4-fluoro-3-hydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate | | Authors: | Kotaka, M, Graeff, R, Zhang, L.H, Lee, H.C, Hao, Q. | | Deposit date: | 2011-08-02 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural Studies of Intermediates Along the Cyclization Pathway of Aplysia Adp-Ribosyl Cyclase.
J.Mol.Biol., 415, 2012
|
|
7X3Y
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle in complex with nAb 9A3 (CVB1-E:9A3) | | Descriptor: | 9A3 heavy chain, 9A3 light chain, VP2, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-03-01 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X35
 
 | | Cryo-EM structure of Coxsackievirus B1 A-particle in complex with nAb 8A10 (CVB1-A:8A10) | | Descriptor: | 8A10 heavy chain, 8A10 light chain, VP2, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-28 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
5WUU
 
 | |
8DW2
 
 | | Cryo-EM structure of SARS-CoV-2 RBD in complex with anti-SARS-CoV-2 DARPin,SR22, and two antibody Fabs, S309 and CR3022 | | Descriptor: | Antibody CR3022 heavy chain, Antibody CR3022 light chain, Antibody S309 heavy chain, ... | | Authors: | Kwon, Y.D, Gorman, J, Kwong, P.D. | | Deposit date: | 2022-07-30 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.11 Å) | | Cite: | A potent and broad neutralization of SARS-CoV-2 variants of concern by DARPins.
Nat.Chem.Biol., 19, 2023
|
|
8DW3
 
 | | Cryo-EM structure of SARS-CoV-2 RBD in complex with anti-SARS-CoV-2 DARPin,SR16m, and two antibody Fabs, S309 and CR3022 | | Descriptor: | Anti-SARS-CoV-2 DARPin SR16m, Antibody S309 light chain, Spike protein S1, ... | | Authors: | Kwon, Y.D, Gorman, J, Kwong, P.D. | | Deposit date: | 2022-07-30 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.26 Å) | | Cite: | A potent and broad neutralization of SARS-CoV-2 variants of concern by DARPins.
Nat.Chem.Biol., 19, 2023
|
|
3KXF
 
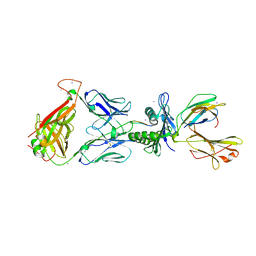 | | Crystal Structure of SB27 TCR in complex with the 'restriction triad' mutant HLA-B*3508-13mer | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-35 alpha chain, ... | | Authors: | Archbold, J.K, Tynan, F.E, Gras, S, Rossjohn, J. | | Deposit date: | 2009-12-03 | | Release date: | 2010-06-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Hard wiring of T cell receptor specificity for the major histocompatibility complex is underpinned by TCR adaptability
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6LGN
 
 | | The atomic structure of varicella zoster virus C-capsid | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | Authors: | Li, S, Zheng, Q. | | Deposit date: | 2019-12-05 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Near-atomic cryo-electron microscopy structures of varicella-zoster virus capsids.
Nat Microbiol, 5, 2020
|
|
6LGL
 
 | | The atomic structure of varicella-zoster virus A-capsid | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | Authors: | Zheng, Q, Li, S. | | Deposit date: | 2019-12-05 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Near-atomic cryo-electron microscopy structures of varicella-zoster virus capsids.
Nat Microbiol, 5, 2020
|
|
