5E6O
 
 | | Crystal structure of C. elegans LGG-2 bound to an AIM/LIR motif | | Descriptor: | Protein lgg-2, TRP-GLU-GLU-LEU | | Authors: | Qi, X, Ren, J.Q, Wu, F, Zhang, H, Feng, W. | | Deposit date: | 2015-10-10 | | Release date: | 2016-01-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of the Differential Function of the Two C. elegans Atg8 Homologs, LGG-1 and LGG-2, in Autophagy
Mol.Cell, 60, 2015
|
|
5E6N
 
 | | Crystal structure of C. elegans LGG-2 | | Descriptor: | Protein lgg-2 | | Authors: | Qi, X, Ren, J.Q, Wu, F, Zhang, H, Feng, W. | | Deposit date: | 2015-10-10 | | Release date: | 2016-01-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structural Basis of the Differential Function of the Two C. elegans Atg8 Homologs, LGG-1 and LGG-2, in Autophagy
Mol.Cell, 60, 2015
|
|
5F21
 
 | | human CD38 in complex with nanobody MU375 | | Descriptor: | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, GLYCEROL, SULFATE ION, ... | | Authors: | Hao, Q, Zhang, H. | | Deposit date: | 2015-12-01 | | Release date: | 2016-06-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Immuno-targeting the multifunctional CD38 using nanobody
Sci Rep, 6, 2016
|
|
5GQQ
 
 | | Structure of ALG-2/HEBP2 Complex | | Descriptor: | CALCIUM ION, CHLORIDE ION, Heme-binding protein 2, ... | | Authors: | Liu, X, Ma, J, Zhang, H, Feng, Y. | | Deposit date: | 2016-08-08 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Functional Study of Apoptosis-linked Gene-2Heme-binding Protein 2 Interactions in HIV-1 Production.
J. Biol. Chem., 291, 2016
|
|
8I23
 
 | | Clostridium thermocellum RNA polymerase transcription open complex with SigI1 and its promoter | | Descriptor: | DNA (80-mer), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Li, J, Zhang, H, Li, D, Feng, Y, Zhu, P. | | Deposit date: | 2023-01-13 | | Release date: | 2023-10-11 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structure of the transcription open complex of distinct sigma I factors.
Nat Commun, 14, 2023
|
|
8I24
 
 | | Clostridium thermocellum RNA polymerase transcription open complex with SigI6 and its promoter | | Descriptor: | DNA (80-mer), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Li, J, Zhang, H, Li, D, Feng, Y, Zhu, P. | | Deposit date: | 2023-01-13 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structure of the transcription open complex of distinct sigma I factors.
Nat Commun, 14, 2023
|
|
4AOJ
 
 | | Human TrkA in complex with the inhibitor AZ-23 | | Descriptor: | 5-chloranyl-N2-[(1S)-1-(5-fluoranylpyridin-2-yl)ethyl]-N4-(3-propan-2-yloxy-1H-pyrazol-5-yl)pyrimidine-2,4-diamine, HIGH AFFINITY NERVE GROWTH FACTOR RECEPTOR, ZINC ION | | Authors: | Wang, T, Lamb, M.L, Block, M.H, Davies, A.M, Han, Y, Hoffmann, E, Ioannidis, S, Josey, J.A, Liu, Z, Lyne, P.D, MacIntyre, T, Mohr, P.J, Omer, C.A, Sjogren, T, Thress, K, Wang, B, Wang, H, Yu, D, Zhang, H. | | Deposit date: | 2012-03-28 | | Release date: | 2012-08-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Discovery of Disubstituted Imidazo[4,5-B]Pyridines and Purines as Potent Trka Inhibitors
Acs Med.Chem.Lett., 3, 2012
|
|
7ETR
 
 | | Crystal structure of SO_1444-SO_1445 complex from Shewanella oneidensis | | Descriptor: | Toxin module of toxin-antitoxin system RelE/StbE family, Transcriptional regulator CopG family | | Authors: | Zhou, J, Zhang, H. | | Deposit date: | 2021-05-13 | | Release date: | 2022-03-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.804 Å) | | Cite: | Insights into the Neutralization and DNA Binding of Toxin-Antitoxin System ParE SO-CopA SO by Structure-Function Studies.
Microorganisms, 9, 2021
|
|
7F61
 
 | |
5YEP
 
 | | Crystal structure of SO_3166-SO_3165 from Shewanella oneidensis | | Descriptor: | Toxin-antitoxin system antidote Mnt family, Toxin-antitoxin system toxin HepN family | | Authors: | Jia, X, Gao, Z.Q, Zhang, H, Dong, Y.H. | | Deposit date: | 2017-09-19 | | Release date: | 2018-03-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-function analyses reveal the molecular architecture and neutralization mechanism of a bacterial HEPN-MNT toxin-antitoxin system.
J. Biol. Chem., 293, 2018
|
|
6AJK
 
 | | Crystal structure of TFB1M and h45 in homo sapiens | | Descriptor: | Dimethyladenosine transferase 1, mitochondrial, RNA (28-MER) | | Authors: | Liu, X, Shen, S, Wu, P, Li, F, Wang, C, Gong, Q, Wu, J, Zhang, H, Shi, Y. | | Deposit date: | 2018-08-28 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Structural insights into dimethylation of 12S rRNA by TFB1M: indispensable role in translation of mitochondrial genes and mitochondrial function.
Nucleic Acids Res., 47, 2019
|
|
5Z6N
 
 | |
6A8R
 
 | | Crystal structure of DUX4 HD2 domain associated with ERG DNA binding site | | Descriptor: | DNA (5'-D(P*AP*AP*TP*CP*TP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*TP*GP*AP*TP*GP*AP*GP*AP*TP*T)-3'), Double homeobox protein 4 | | Authors: | Dong, X, Zhang, H, Cheng, N, Meng, G. | | Deposit date: | 2018-07-10 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | DUX4HD2-DNAERGstructure reveals new insight into DUX4-Responsive-Element.
Leukemia, 33, 2019
|
|
6AAX
 
 | | Crystal structure of TFB1M and h45 with SAM in homo sapiens | | Descriptor: | DI(HYDROXYETHYL)ETHER, Dimethyladenosine transferase 1, mitochondrial, ... | | Authors: | Liu, X, Shen, S, Wu, P, Li, F, Gong, Q, Wu, J, Zhang, H, Shi, Y. | | Deposit date: | 2018-07-19 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.994 Å) | | Cite: | Structural insights into dimethylation of 12S rRNA by TFB1M: indispensable role in translation of mitochondrial genes and mitochondrial function.
Nucleic Acids Res., 47, 2019
|
|
3ES9
 
 | | NADPH-Cytochrome P450 Reductase in an Open Conformation | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Hamdane, D, Xia, C, Im, S.-C, Zhang, H, Kim, J.-J, Waskell, L. | | Deposit date: | 2008-10-05 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure and function of an NADPH-cytochrome P450 oxidoreductase in an open conformation capable of reducing cytochrome P450
J.Biol.Chem., 284, 2009
|
|
1AE2
 
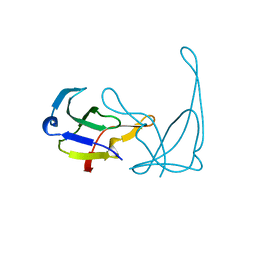 | | MUTANT L32R OF GENE V PROTEIN (SINGLE-STRANDED DNA BINDING PROTEIN) | | Descriptor: | GENE V PROTEIN | | Authors: | Su, S, Gao, Y.-G, Zhang, H, Terwilliger, T.C, Wang, A.H.-J. | | Deposit date: | 1997-03-04 | | Release date: | 1997-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analyses of the stability and function of three surface mutants (R82C, K69H, and L32R) of the gene V protein from Ff phage by X-ray crystallography.
Protein Sci., 6, 1997
|
|
1AE3
 
 | | MUTANT R82C OF GENE V PROTEIN (SINGLE-STRANDED DNA BINDING PROTEIN) | | Descriptor: | GENE V PROTEIN | | Authors: | Su, S, Gao, Y.-G, Zhang, H, Terwilliger, T.C, Wang, A.H.-J. | | Deposit date: | 1997-03-04 | | Release date: | 1997-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analyses of the stability and function of three surface mutants (R82C, K69H, and L32R) of the gene V protein from Ff phage by X-ray crystallography.
Protein Sci., 6, 1997
|
|
8I2H
 
 | | Follicle stimulating hormone receptor | | Descriptor: | Follicle-stimulating hormone receptor | | Authors: | Duan, J, Xu, P, Yang, J, Ji, Y, Zhang, H, Mao, C, Luan, X, Jiang, Y, Zhang, Y, Zhang, S, Xu, H.E. | | Deposit date: | 2023-01-14 | | Release date: | 2023-03-22 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Mechanism of hormone and allosteric agonist mediated activation of follicle stimulating hormone receptor.
Nat Commun, 14, 2023
|
|
8I2G
 
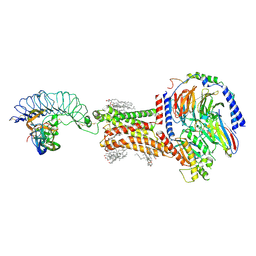 | | FSHR-Follicle stimulating hormone-compound 716340-Gs complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Duan, J, Xu, P, Yang, J, Ji, Y, Zhang, H, Mao, C, Luan, X, Jiang, Y, Zhang, Y, Zhang, S, Xu, H.E. | | Deposit date: | 2023-01-14 | | Release date: | 2023-03-29 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of hormone and allosteric agonist mediated activation of follicle stimulating hormone receptor.
Nat Commun, 14, 2023
|
|
1BVY
 
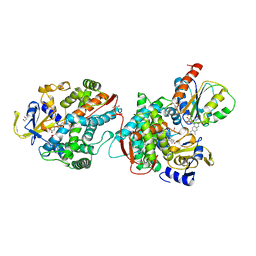 | | COMPLEX OF THE HEME AND FMN-BINDING DOMAINS OF THE CYTOCHROME P450(BM-3) | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, PROTEIN (CYTOCHROME P450 BM-3), ... | | Authors: | Sevrioukova, I.F, Li, H, Zhang, H, Peterson, J.A, Poulos, T.L. | | Deposit date: | 1998-09-21 | | Release date: | 1999-02-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure of a cytochrome P450-redox partner electron-transfer complex.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
7XTB
 
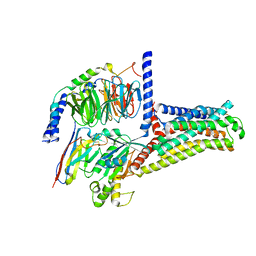 | | Serotonin 6 (5-HT6) receptor-Gs-Nb35 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Huang, S, Xu, P, Shen, D.D, Simon, I.A, Mao, C, Tan, Y, Zhang, H, Harpsoe, K, Li, H, Zhang, Y, You, C, Yu, X, Jiang, Y, Zhang, Y, Gloriam, D.E, Xu, H.E. | | Deposit date: | 2022-05-16 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | GPCRs steer G i and G s selectivity via TM5-TM6 switches as revealed by structures of serotonin receptors.
Mol.Cell, 82, 2022
|
|
7XTA
 
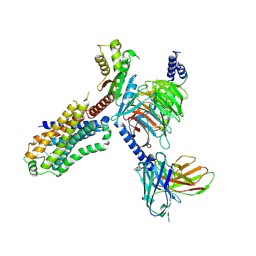 | | Serotonin 4 (5-HT4) receptor-Gi-scFv16 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Huang, S, Xu, P, Shen, D.D, Simon, I.A, Mao, C, Tan, Y, Zhang, H, Harpsoe, K, Li, H, Zhang, Y, You, C, Yu, X, Jiang, Y, Zhang, Y, Gloriam, D.E, Xu, H.E. | | Deposit date: | 2022-05-16 | | Release date: | 2022-07-27 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | GPCRs steer G i and G s selectivity via TM5-TM6 switches as revealed by structures of serotonin receptors.
Mol.Cell, 82, 2022
|
|
7XTC
 
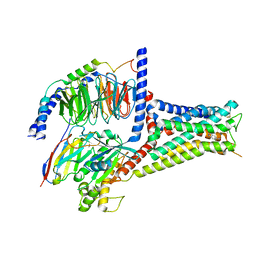 | | Serotonin 7 (5-HT7) receptor-Gs-Nb35 complex | | Descriptor: | 3-(2-azanylethyl)-1H-indole-5-carboxamide, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Huang, S, Xu, P, Shen, D.D, Simon, I.A, Mao, C, Tan, Y, Zhang, H, Harpsoe, K, Li, H, Zhang, Y, You, C, Yu, X, Jiang, Y, Zhang, Y, Gloriam, D.E, Xu, H.E. | | Deposit date: | 2022-05-16 | | Release date: | 2022-07-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | GPCRs steer G i and G s selectivity via TM5-TM6 switches as revealed by structures of serotonin receptors.
Mol.Cell, 82, 2022
|
|
7XT8
 
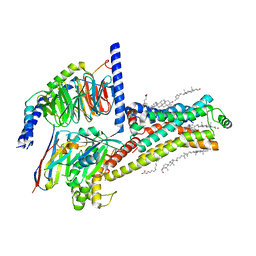 | | Serotonin 4 (5-HT4) receptor-Gs-Nb35 complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Huang, S, Xu, P, Shen, D.D, Simon, I.A, Mao, C, Tan, Y, Zhang, H, Harpsoe, K, Li, H, Zhang, Y, You, C, Yu, X, Jiang, Y, Zhang, Y, Gloriam, D.E, Xu, H.E. | | Deposit date: | 2022-05-16 | | Release date: | 2022-07-27 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | GPCRs steer G i and G s selectivity via TM5-TM6 switches as revealed by structures of serotonin receptors.
Mol.Cell, 82, 2022
|
|
7XT9
 
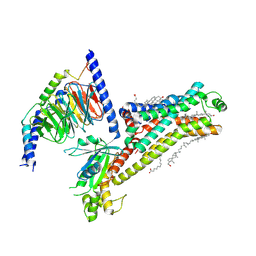 | | Serotonin 4 (5-HT4) receptor-Gs complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Huang, S, Xu, P, Shen, D.D, Simon, I.A, Mao, C, Tan, Y, Zhang, H, Harpsoe, K, Li, H, Zhang, Y, You, C, Yu, X, Jiang, Y, Zhang, Y, Gloriam, D.E, Xu, H.E. | | Deposit date: | 2022-05-16 | | Release date: | 2022-07-27 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | GPCRs steer G i and G s selectivity via TM5-TM6 switches as revealed by structures of serotonin receptors.
Mol.Cell, 82, 2022
|
|
