3NU5
 
 | | Crystal Structure of HIV-1 Protease Mutant I50V with Antiviral Drug Amprenavir | | Descriptor: | ACETATE ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Wang, Y.-F, Shen, C.H, Weber, I.T. | | Deposit date: | 2010-07-06 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Amprenavir complexes with HIV-1 protease and its drug-resistant mutants altering hydrophobic clusters.
Febs J., 277, 2010
|
|
3NYF
 
 | |
4U8W
 
 | | HIV-1 wild Type protease with GRL-050-10A (a Gem-difluoro-bis-Tetrahydrofuran as P2-Ligand) | | Descriptor: | (3R,3aS,6aS)-4,4-difluorohexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{[(4-aminophenyl)sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2014-08-05 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Design of gem-Difluoro-bis-Tetrahydrofuran as P2 Ligand for HIV-1 Protease Inhibitors to Improve Brain Penetration: Synthesis, X-ray Studies, and Biological Evaluation.
Chemmedchem, 10, 2015
|
|
5T2E
 
 | |
5T2Z
 
 | | Crystal Structure of Multi-drug Resistant HIV-1 Protease PR-S17 in Complex with Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | Authors: | Agniswamy, J, Weber, I.T. | | Deposit date: | 2016-08-24 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Studies of a Rationally Selected Multi-Drug Resistant HIV-1 Protease Reveal Synergistic Effect of Distal Mutations on Flap Dynamics.
PLoS ONE, 11, 2016
|
|
3IL8
 
 | | CRYSTAL STRUCTURE OF INTERLEUKIN 8: SYMBIOSIS OF NMR AND CRYSTALLOGRAPHY | | Descriptor: | INTERLEUKIN-8 | | Authors: | Baldwin, E.T, Weber, I.T, St Charles, R, Xuan, J.-C, Appella, E, Yamada, M, Matsushima, K, Edwards, B.F.P, Clore, G.M, Gronenborn, A.M, Wlodawer, A. | | Deposit date: | 1990-12-07 | | Release date: | 1992-10-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of interleukin 8: symbiosis of NMR and crystallography.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
3LJP
 
 | | Crystal structure of choline oxidase V464A mutant | | Descriptor: | Choline oxidase, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE | | Authors: | Finnegan, S, Agniswamy, J, Weber, I.T, Giovanni, G. | | Deposit date: | 2010-01-26 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Role of valine 464 in the flavin oxidation reaction catalyzed by choline oxidase.
Biochemistry, 49, 2010
|
|
5JG1
 
 | | HIV-1 wild Type protease with GRL-031-14A (a Adamantane P1-Ligand with tetrahydropyrano-tetrahydrofuran in P2 and isobutylamine in P1') | | Descriptor: | (3R,3aS,7aR)-hexahydro-4H-furo[2,3-b]pyran-3-yl {(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-[(3R,5R,7R)-tricyclo[3.3.1.1~3,7~]decan-1-yl]butan-2-yl}carbamate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2016-04-19 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Probing Lipophilic Adamantyl Group as the P1-Ligand for HIV-1 Protease Inhibitors: Design, Synthesis, Protein X-ray Structural Studies, and Biological Evaluation.
J.Med.Chem., 59, 2016
|
|
3JVW
 
 | | HIV-1 Protease Mutant G86A with symmetric inhibitor DMP323 | | Descriptor: | Gag-Pol polyprotein, [4-R-(-4-ALPHA,5-ALPHA,6-BETA,7-BETA)]-HEXAHYDRO-5,6-BIS(HYDROXY)-[1,3-BIS([4-HYDROXYMETHYL-PHENYL]METHYL)-4,7-BIS(PHEN YLMETHYL)]-2H-1,3-DIAZEPINONE | | Authors: | Tie, Y, Weber, I.T. | | Deposit date: | 2009-09-17 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Highly conserved glycine 86 and arginine 87 residues contribute differently to the structure and activity of the mature HIV-1 protease
Proteins, 78, 2009
|
|
3JW2
 
 | | HIV-1 Protease Mutant G86S with DARUNAVIR | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, Gag-Pol polyprotein, ... | | Authors: | Tie, Y, Weber, I.T. | | Deposit date: | 2009-09-17 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Highly conserved glycine 86 and arginine 87 residues contribute differently to the structure and activity of the mature HIV-1 protease
Proteins, 78, 2009
|
|
3JVY
 
 | | HIV-1 Protease Mutant G86A with DARUNAVIR | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, Gag-Pol polyprotein, ... | | Authors: | Tie, Y, Weber, I.T. | | Deposit date: | 2009-09-17 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Highly conserved glycine 86 and arginine 87 residues contribute differently to the structure and activity of the mature HIV-1 protease
Proteins, 78, 2009
|
|
3MWS
 
 | | Crystal Structure of Group N HIV-1 Protease | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, HIV-1 Protease | | Authors: | Sayer, J.M, Agniswamy, J, Weber, I.T, Louis, J.M. | | Deposit date: | 2010-05-06 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Autocatalytic maturation, physical/chemical properties, and crystal structure of group N HIV-1 protease: relevance to drug resistance.
Protein Sci., 19, 2010
|
|
7RDF
 
 | | Crystal structure of Pseudomonas aeruginosa D-Arginine Dehydrogenase Y249F co-crystallized in the presence of D-arginine | | Descriptor: | 6-HYDROXY-FLAVIN-ADENINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, FAD-dependent catabolic D-arginine dehydrogenase DauA, ... | | Authors: | Reis, R.A.G, Iyer, A, Agniswamy, A, Weber, I.T, Gadda, G. | | Deposit date: | 2021-07-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Discovery of a new flavin N5-adduct in a tyrosine to phenylalanine variant of d-Arginine dehydrogenase.
Arch.Biochem.Biophys., 715, 2021
|
|
2IEN
 
 | | Crystal structure analysis of HIV-1 protease with a potent non-peptide inhibitor (UIC-94017) | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Tie, Y, Boross, P.I, Wang, Y.F, Gaddis, L, Manna, D, Hussain, A.K, Leshchenko, S, Ghosh, A.K, Louis, J.M, Harrison, R.W, Weber, I.T. | | Deposit date: | 2006-09-19 | | Release date: | 2006-10-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High Resolution Crystal Structures of HIV-1 Protease with a Potent Non-Peptide Inhibitor (Uic-94017) Active Against Multi-Drug-Resistant Clinical Strains.
J.Mol.Biol., 338, 2004
|
|
5ULT
 
 | | HIV-1 wild Type protease with GRL-100-13A (a Crown-like Oxotricyclic Core as the P2-Ligand with the sulfonamide isostere as the P2' group) | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, Protease, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Design and Development of Highly Potent HIV-1 Protease Inhibitors with a Crown-Like Oxotricyclic Core as the P2-Ligand To Combat Multidrug-Resistant HIV Variants.
J. Med. Chem., 60, 2017
|
|
5UPZ
 
 | | HIV-1 wild Type protease with GRL-0518A , an isophthalamide-derived P2-P3 ligand with the para-hydoxymethyl sulfonamide isostere as the P2' group | | Descriptor: | CHLORIDE ION, GLYCEROL, N~3~-{(2S,3R)-3-hydroxy-4-[{[4-(hydroxymethyl)phenyl]sulfonyl}(2-methylpropyl)amino]-1-phenylbutan-2-yl}-N~1~-methyl-N~1~-[(4-methyl-1,3-oxazol-2-yl)methyl]benzene-1,3-dicarboxamide, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2017-02-05 | | Release date: | 2017-05-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Design of novel HIV-1 protease inhibitors incorporating isophthalamide-derived P2-P3 ligands: Synthesis, biological evaluation and X-ray structural studies of inhibitor-HIV-1 protease complex.
Bioorg. Med. Chem., 25, 2017
|
|
5UOV
 
 | | HIV-1 wild Type protease with GRL-1118A , an isophthalamide-derived P2-P3 ligand with the sulfonamide isostere as the P2' group | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2017-02-01 | | Release date: | 2017-05-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Design of novel HIV-1 protease inhibitors incorporating isophthalamide-derived P2-P3 ligands: Synthesis, biological evaluation and X-ray structural studies of inhibitor-HIV-1 protease complex.
Bioorg. Med. Chem., 25, 2017
|
|
5WLO
 
 | | a novel 13-ring macrocyclic HIV-1 protease inhibitors involving the P1'-P2' ligands | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-3-hydroxy-4-[(7E)-13-methoxy-1,1-dioxo-1,4,5,6,9,11-hexahydro-10,1lambda~6~,2-benzoxathiazacyclotridecin-2(3H)-yl]-1-phenylbutan-2-yl}carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2017-07-27 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Design, synthesis, X-ray studies, and biological evaluation of novel macrocyclic HIV-1 protease inhibitors involving the P1'-P2' ligands.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
3BVB
 
 | | Cystal structure of HIV-1 Active Site Mutant D25N and inhibitor Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Liu, F, Weber, I.T. | | Deposit date: | 2008-01-05 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Effect of the Active Site D25N Mutation on the Structure, Stability, and Ligand Binding of the Mature HIV-1 Protease.
J.Biol.Chem., 283, 2008
|
|
3BVA
 
 | | Cystal structure of HIV-1 Active Site Mutant D25N and p2-NC analog inhibitor | | Descriptor: | GLYCEROL, N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide, Protease (Retropepsin) | | Authors: | Liu, F, Weber, I.T. | | Deposit date: | 2008-01-05 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of the Active Site D25N Mutation on the Structure, Stability, and Ligand Binding of the Mature HIV-1 Protease.
J.Biol.Chem., 283, 2008
|
|
7TO5
 
 | | HIV-1 wild type protease with GRL-05816A, with C-4 substituted cyclohexane-fused bis-tetrahydrofuran (Chf-THF) derivatives as P2-ligand [diastereomer 1] | | Descriptor: | (1R,3aS,4S,6S,7aR)-octahydro-1,6-epoxy-2-benzofuran-4-yl [(2S,3R)-4-{[2-(cyclopropylamino)-1,3-benzothiazole-6-sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Ghosh, A.K, Weber, I.T. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Design, Synthesis and X-Ray Structural Studies of Potent HIV-1 Protease Inhibitors Containing C-4 Substituted Tricyclic Hexahydro-Furofuran Derivatives as P2 Ligands.
Chemmedchem, 17, 2022
|
|
7TO6
 
 | | HIV-1 wild type protease with GRL-01717A, with C-4 substituted cyclohexane-fused bis-tetrahydrofuran (Chf-THF) derivatives as P2-ligand [diastereomer 2] | | Descriptor: | (1S,3aR,4R,6R,7aS)-octahydro-1,6-epoxy-2-benzofuran-4-yl [(2S,3R)-4-{[2-(cyclopropylamino)-1,3-benzothiazole-6-sulfonyl](2-methylpropyl)amino}-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Ghosh, A.K, Weber, I.T. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Design, Synthesis and X-Ray Structural Studies of Potent HIV-1 Protease Inhibitors Containing C-4 Substituted Tricyclic Hexahydro-Furofuran Derivatives as P2 Ligands.
Chemmedchem, 17, 2022
|
|
8F0F
 
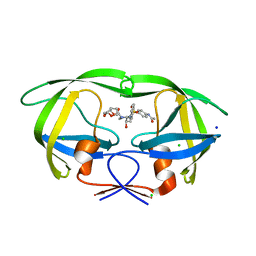 | | HIV-1 wild type protease with GRL-110-19A, a chloroacetamide derivative based on Darunavir as P2' group | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{[4-(2-chloroacetamido)benzene-1-sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Ghosh, A.K, Weber, I.T. | | Deposit date: | 2022-11-02 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Evaluation of darunavir-derived HIV-1 protease inhibitors incorporating P2' amide-derivatives: Synthesis, biological evaluation and structural studies.
Bioorg.Med.Chem.Lett., 83, 2023
|
|
8FUJ
 
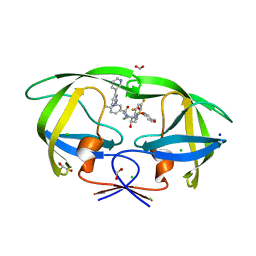 | | HIV-1 wild type protease with GRL-03419A, with N-(2,5-dimethylphenyl)-4-(pyridin-3-yl)pyrimidin-2-amine as P2-P3 group and 3,5-difluorophenylmethyl as the P1 group | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Ghosh, A.K, Weber, I.T. | | Deposit date: | 2023-01-17 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Exploration of imatinib and nilotinib-derived templates as the P2-Ligand for HIV-1 protease inhibitors: Design, synthesis, protein X-ray structural studies, and biological evaluation.
Eur.J.Med.Chem., 255, 2023
|
|
8FUI
 
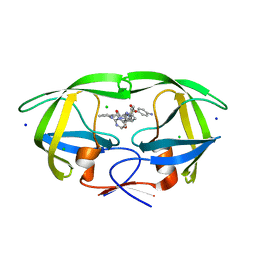 | | HIV-1 wild type protease with GRL-02519A, with N-(2,5-dimethylphenyl)-4-(pyridin-3-yl)pyrimidin-2-amine as P2-P3 group | | Descriptor: | ACETATE ION, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Wang, Y.-F, Wong-Sam, A.E, Ghosh, A.K, Weber, I.T. | | Deposit date: | 2023-01-17 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Exploration of imatinib and nilotinib-derived templates as the P2-Ligand for HIV-1 protease inhibitors: Design, synthesis, protein X-ray structural studies, and biological evaluation.
Eur.J.Med.Chem., 255, 2023
|
|
