2PK8
 
 | | Crystal structure of an uncharacterized protein PF0899 from Pyrococcus furiosus | | Descriptor: | GOLD (I) CYANIDE ION, Uncharacterized protein PF0899 | | Authors: | Liu, Z.J, Tempel, W, Chen, L, Shah, A, Lee, D, Clancy-Kelley, L.L, Dillard, B.D, Rose, J.P, Sugar, F.J, Jenny Jr, F.E, Lee, H.S, Izumi, M, Shah, C, Poole III, F.L, Adams, M.W.W, Richardson, J.S, Richardson, D.C, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2007-04-17 | | Release date: | 2007-05-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the hypothetical protein PF0899 from Pyrococcus furiosus at 1.85 A resolution.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
6BX8
 
 | | Human Mesotrypsin (PRSS3) Complexed with Tissue Factor Pathway Inhibitor Variant (TFPI1-KD1-K15R-I17C-I34C) | | Descriptor: | SULFATE ION, Tissue factor pathway inhibitor, Trypsin-3 | | Authors: | Coban, M, Sankaran, B, Cohen, I, Hockla, A, Papo, N, Radisky, E.S. | | Deposit date: | 2017-12-18 | | Release date: | 2019-02-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Disulfide engineering of human Kunitz-type serine protease inhibitors enhances proteolytic stability and target affinity toward mesotrypsin.
J. Biol. Chem., 294, 2019
|
|
9QOF
 
 | | E.coli seryl-tRNA synthetase (Arm deletion mutant) bound to sulphamoyl seryl-adenylate analogue | | Descriptor: | 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Cusack, S, Belrhali, H, Price, S, Leberman, R. | | Deposit date: | 2025-03-26 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Seryl-tRNA synthetase from Escherichia coli,implication of its N-terminal domain in aminoacylation activity and specificity.
Nucleic Acids Res, 22, 1994
|
|
7BJ3
 
 | | ScpA from Streptococcus pyogenes, S512A active site mutant | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, C5a peptidase, CALCIUM ION, ... | | Authors: | Kagawa, T.F, O'Connell, M.R, Cooney, J.C. | | Deposit date: | 2021-01-13 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Enzyme kinetic and binding studies identify determinants of specificity for the immunomodulatory enzyme ScpA, a C5a inactivating bacterial protease.
Comput Struct Biotechnol J, 19, 2021
|
|
7BBM
 
 | | Mutant nitrobindin M75L/H76L/Q96C/M148L (NB4H) from Arabidopsis thaliana with cofactor MnPPIX | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE PROTOPORPHYRIN IX, UPF0678 fatty acid-binding protein-like protein At1g79260 | | Authors: | Minges, A, Sauer, D.F, Wittwer, M, Markel, U, Spiertz, M, Schiffels, J, Davari, M.D, Okuda, J, Schwaneberg, U, Groth, G. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Chemogenetic engineering of nitrobindin toward an artificial epoxygenase
Catalysis Science And Technology, 2021
|
|
6XA9
 
 | | SARS CoV-2 PLpro in complex with ISG15 C-terminal domain propargylamide | | Descriptor: | GLYCEROL, ISG15 CTD-propargylamide, Non-structural protein 3, ... | | Authors: | Klemm, T, Calleja, D.J, Richardson, L.W, Lechtenberg, B.C, Komander, D. | | Deposit date: | 2020-06-04 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2.
Embo J., 39, 2020
|
|
5KAF
 
 | | RT XFEL structure of Photosystem II in the dark state at 3.0 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Fuller, F, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Waterman, D.G, Evans, G, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-06-01 | | Release date: | 2016-11-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.00001 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
5KAI
 
 | | NH3-bound RT XFEL structure of Photosystem II 500 ms after the 2nd illumination (2F) at 2.8 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Fuller, F, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Waterman, D.G, Evans, G, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-06-01 | | Release date: | 2016-11-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.80000925 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
6MF2
 
 | | Improved Model of Human Coagulation Factor VIII | | Descriptor: | CALCIUM ION, COPPER (I) ION, Coagulation factor VIII, ... | | Authors: | Smith, I.W, Spiegel, P.C. | | Deposit date: | 2018-09-08 | | Release date: | 2019-09-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.609364 Å) | | Cite: | The 3.2 angstrom structure of a bioengineered variant of blood coagulation factor VIII indicates two conformations of the C2 domain.
J.Thromb.Haemost., 18, 2020
|
|
6MF0
 
 | | Crystal Structure Determination of Human/Porcine Chimera Coagulation Factor VIII | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, COPPER (I) ION, ... | | Authors: | Smith, I.W, Spiegel, P.C. | | Deposit date: | 2018-09-07 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.19999886 Å) | | Cite: | The 3.2 angstrom structure of a bioengineered variant of blood coagulation factor VIII indicates two conformations of the C2 domain.
J.Thromb.Haemost., 18, 2020
|
|
7JMC
 
 | | Sheep Connexin-50 at 2.5 angstroms resolution, Lipid Class 3 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-8 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.D, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-07-31 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
7JIZ
 
 | |
7JY2
 
 | | Z-DNA joint X-ray/Neutron | | Descriptor: | Chains: A,B | | Authors: | Harp, J.M, Coates, L, Egli, M. | | Deposit date: | 2020-08-28 | | Release date: | 2021-04-28 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.5 Å), X-RAY DIFFRACTION | | Cite: | Water structure around a left-handed Z-DNA fragment analyzed by cryo neutron crystallography.
Nucleic Acids Res., 49, 2021
|
|
7C7J
 
 | |
7C7I
 
 | |
9ONO
 
 | | Fe-bound B. pseudomallei rubrerythrin | | Descriptor: | FE (III) ION, Rubrerythrin | | Authors: | Budziszewski, G.R, Snell, M.E, Monteiro, D.C.F, Lynch, M.L, Bowman, S.E.J. | | Deposit date: | 2025-05-15 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Burkholderia pseudomallei rubrerythrin promiscuously binds metals in a structurally pre-formed bimetallic binding site.
Biorxiv, 2025
|
|
9ONN
 
 | | Co-bound B. pseudomallei Rubrerythrin | | Descriptor: | COBALT (II) ION, Rubrerythrin | | Authors: | Budziszewski, G.R, Snell, M.E, Monteiro, D.C.F, Lynch, M.L, Bowman, S.E.J. | | Deposit date: | 2025-05-15 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Burkholderia pseudomallei rubrerythrin promiscuously binds metals in a structurally pre-formed bimetallic binding site.
Biorxiv, 2025
|
|
9ONM
 
 | | near-apo rubrerythrin | | Descriptor: | DI(HYDROXYETHYL)ETHER, FE (III) ION, Rubrerythrin | | Authors: | Budziszewski, G.R, Snell, M.E, Monteiro, D.C.F, Lynch, M.L, Bowman, S.E.J. | | Deposit date: | 2025-05-15 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Burkholderia pseudomallei rubrerythrin promiscuously binds metals in a structurally pre-formed bimetallic binding site.
Biorxiv, 2025
|
|
9ONR
 
 | | rubrerythrin- E53A E124A | | Descriptor: | DI(HYDROXYETHYL)ETHER, Rubrerythrin, THIOCYANATE ION | | Authors: | Budziszewski, G.R, Snell, M.E, Monteiro, D.C.F, Lynch, M.L, Bowman, S.E.J. | | Deposit date: | 2025-05-15 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Burkholderia pseudomallei rubrerythrin promiscuously binds metals in a structurally pre-formed bimetallic binding site.
Biorxiv, 2025
|
|
9ONQ
 
 | | Mn-bound B. pseudomallei rubrerythrin | | Descriptor: | MANGANESE (II) ION, Rubrerythrin | | Authors: | Budziszewski, G.R, Snell, M.E, Monteiro, D.C.F, Lynch, M.L, Bowman, S.E.J. | | Deposit date: | 2025-05-15 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Burkholderia pseudomallei rubrerythrin promiscuously binds metals in a structurally pre-formed bimetallic binding site.
Biorxiv, 2025
|
|
8FSD
 
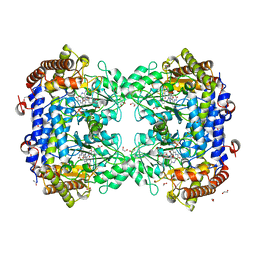 | | P130R mutant of soybean SHMT8 in complex with PLP-glycine and formylTHF | | Descriptor: | 1,2-ETHANEDIOL, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, ... | | Authors: | Beamer, L.J, Korasick, D.A. | | Deposit date: | 2023-01-09 | | Release date: | 2023-10-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural and functional analysis of two SHMT8 variants associated with soybean cyst nematode resistance.
Febs J., 291, 2024
|
|
8G8F
 
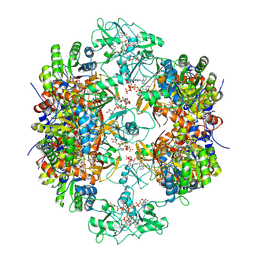 | | Human IMPDH2 mutant - L245P, treated with ATP, IMP, and NAD+; extended filament segment reconstruction | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase 2, ... | | Authors: | O'Neill, A.G, Kollman, J.M. | | Deposit date: | 2023-02-17 | | Release date: | 2023-04-19 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Neurodevelopmental disorder mutations in the purine biosynthetic enzyme IMPDH2 disrupt its allosteric regulation.
J.Biol.Chem., 299, 2023
|
|
8G9B
 
 | | Human IMPDH2 mutant - L245P, treated with GTP, ATP, IMP, and NAD+; compressed filament segment reconstruction | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | O'Neill, A.G, Kollman, J.M. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-19 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Neurodevelopmental disorder mutations in the purine biosynthetic enzyme IMPDH2 disrupt its allosteric regulation.
J.Biol.Chem., 299, 2023
|
|
5HLL
 
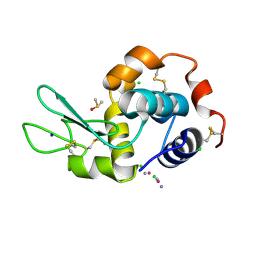 | |
5I6U
 
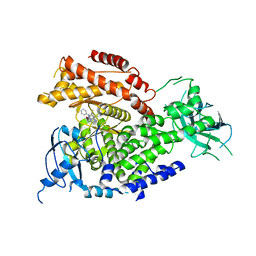 | | The crystal structure of PI3Kdelta with compound 32 | | Descriptor: | 2-[(1S)-1-({6-amino-5-[(1H-pyrazol-4-yl)ethynyl]pyrimidin-4-yl}amino)ethyl]-5-chloro-3-phenylquinazolin-4(3H)-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Somoza, J.R, Villasenor, A.G. | | Deposit date: | 2016-02-16 | | Release date: | 2017-02-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.842 Å) | | Cite: | The crystal structure of PI3Kdelta with compound 32
To Be Published
|
|
