9BAP
 
 | | CryoEM structure of Apo-DIM2 | | Descriptor: | DNA (cytosine-5-)-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Song, J, Shao, Z. | | Deposit date: | 2024-04-04 | | Release date: | 2024-07-24 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Multi-layered heterochromatin interaction as a switch for DIM2-mediated DNA methylation.
Nat Commun, 15, 2024
|
|
9BAZ
 
 | | CryoEM structure of DIM2-HP1 complex | | Descriptor: | DNA (cytosine-5-)-methyltransferase, Heterochromatin protein one, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Song, J, Shao, Z. | | Deposit date: | 2024-04-05 | | Release date: | 2024-07-24 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Multi-layered heterochromatin interaction as a switch for DIM2-mediated DNA methylation.
Nat Commun, 15, 2024
|
|
9BAQ
 
 | | CryoEM structure of DIM2-HP1-H3K9me3-DNA complex | | Descriptor: | DNA (5'-D(*AP*CP*TP*AP*CP*T)-R(P*(PYO))-D(P*CP*TP*CP*CP*TP*CP*CP*TP*AP*CP*T)-3'), DNA (5'-D(*AP*GP*TP*AP*GP*GP*AP*GP*GP*AP*GP*GP*AP*GP*TP*AP*GP*T)-3'), DNA (cytosine-5-)-methyltransferase, ... | | Authors: | Song, J, Shao, Z. | | Deposit date: | 2024-04-04 | | Release date: | 2024-07-24 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Multi-layered heterochromatin interaction as a switch for DIM2-mediated DNA methylation.
Nat Commun, 15, 2024
|
|
9L8G
 
 | | R115A mutant of Ferredoxin-NADP+ reductase from maize root - Oxidized form, low X-ray dose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase, ... | | Authors: | Uenaka, M, Ohnishi, Y, Tanaka, H, Kurisu, G. | | Deposit date: | 2024-12-27 | | Release date: | 2025-01-29 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Redox-dependent hydrogen-bond network rearrangement of ferredoxin-NADP + reductase revealed by high-resolution X-ray and neutron crystallography.
Acta Crystallogr.,Sect.F, 81, 2025
|
|
8RPC
 
 | | Crystal structure of PfCLK3 with TCMDC-135051 | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-[5-(diethylaminomethyl)-2-methoxy-phenyl]-1~{H}-pyrrolo[2,3-b]pyridin-4-yl]-2-propan-2-yl-benzoic acid, GLYCEROL, ... | | Authors: | Yelland, T.S, Benazir, A, Hole, A. | | Deposit date: | 2024-01-15 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.079 Å) | | Cite: | Targeting Pf CLK3 with Covalent Inhibitors: A Novel Strategy for Malaria Treatment.
J.Med.Chem., 67, 2024
|
|
8QHF
 
 | | Corynebacterium glutamicum mycoloyltransferase C acyl-enzyme intermediate | | Descriptor: | (2~{S},3~{R})-2-pentyloctane-1,3-diol, CHLORIDE ION, Cmt1, ... | | Authors: | Li de la Sierra-Gallay, I, Lesur, E. | | Deposit date: | 2023-09-08 | | Release date: | 2025-02-12 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Synthetic mycolates derivatives to decipher protein mycoloylation, a unique post-translational modification in bacteria.
J.Biol.Chem., 301, 2025
|
|
4X1D
 
 | | Ytterbium-bound human serum transferrin | | Descriptor: | GLYCEROL, MALONATE ION, Serotransferrin, ... | | Authors: | Wang, M, Zhang, H, Sun, H. | | Deposit date: | 2014-11-24 | | Release date: | 2015-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | "Anion clamp" allows flexible protein to impose coordination geometry on metal ions
Chem.Commun.(Camb.), 51, 2015
|
|
4X1B
 
 | | Human serum transferrin with ferric ion bound at the C-lobe only | | Descriptor: | FE (III) ION, GLYCEROL, MALONATE ION, ... | | Authors: | Wang, M, Zhang, H, Sun, H. | | Deposit date: | 2014-11-24 | | Release date: | 2015-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | "Anion clamp" allows flexible protein to impose coordination geometry on metal ions
Chem.Commun.(Camb.), 51, 2015
|
|
8BA7
 
 | |
8BA8
 
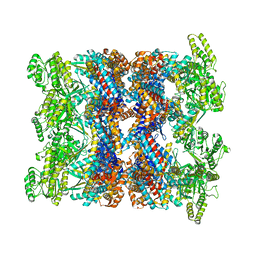 | | CryoEM structure of GroEL-ADP.BeF3-Rubisco. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chaperonin GroEL, ... | | Authors: | Gardner, S, Saibil, H.R. | | Deposit date: | 2022-10-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of substrate progression through the bacterial chaperonin cycle.
Proc Natl Acad Sci U S A, 120, 2023
|
|
8BA9
 
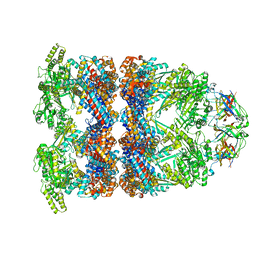 | | CryoEM structure of GroEL-GroES-ADP.AlF3-Rubisco. | | Descriptor: | 60 kDa chaperonin, ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, ... | | Authors: | Gardner, S, Saibil, H.R. | | Deposit date: | 2022-10-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of substrate progression through the bacterial chaperonin cycle.
Proc Natl Acad Sci U S A, 120, 2023
|
|
8BAA
 
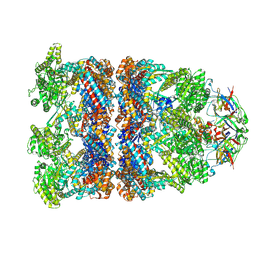 | |
1JCA
 
 | | Non-standard Design of Unstable Insulin Analogues with Enhanced Activity | | Descriptor: | ZINC ION, insulin a, insulin b | | Authors: | Weiss, M.A, Wan, Z, Zhao, M, Chu, Y.-C, Nakagawa, S.H, Burke, G.T, Jia, W, Hellmich, R, Katsoyannis, P.G. | | Deposit date: | 2001-06-08 | | Release date: | 2001-06-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Non-standard insulin design: structure-activity relationships at the periphery of the insulin receptor.
J.Mol.Biol., 315, 2002
|
|
6DDS
 
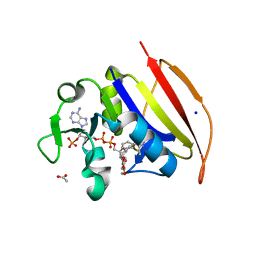 | | Mycobacterium tuberculosis Dihydrofolate Reductase complexed with beta-NADPH and 4-[3-[3-[2,4-bis(azanyl)-6-ethyl-pyrimidin-5-yl]prop-2-ynyl]-5-methoxy-phenyl]benzoic acid | | Descriptor: | 4-[3-[3-[2,4-bis(azanyl)-6-ethyl-pyrimidin-5-yl]prop-2-ynyl]-5-methoxy-phenyl]benzoic acid, ACETATE ION, Dihydrofolate reductase, ... | | Authors: | Hajian, B, Wright, D, Scocchera, E. | | Deposit date: | 2018-05-10 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Drugging the Folate Pathway in Mycobacterium tuberculosis: The Role of Multi-targeting Agents.
Cell Chem Biol, 26, 2019
|
|
6DE4
 
 | | Homo sapiens dihydrofolate reductase complexed with beta-NADPH and 3'-[(2R)-4-(2,4-diamino-6-ethylphenyl)but-3-yn-2-yl]-5'-methoxy-[1,1'-biphenyl]-4-carboxylic acid | | Descriptor: | 3'-[(2R)-4-(2,4-diamino-6-ethylpyrimidin-5-yl)but-3-yn-2-yl]-5'-methoxy[1,1'-biphenyl]-4-carboxylic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hajian, B, Wright, D. | | Deposit date: | 2018-05-11 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.411 Å) | | Cite: | Drugging the Folate Pathway in Mycobacterium tuberculosis: The Role of Multi-targeting Agents.
Cell Chem Biol, 26, 2019
|
|
6ND2
 
 | | Staphylococcus aureus Dihydrofolate Reductase complexed with NADPH and 6-ETHYL-5-[(3R)-3-[2-METHOXY-5-(PYRIDIN-4-YL)PHENYL]BUT-1-YN-1-YL]PYRIMIDINE-2,4-DIAMINE (UCP1063) | | Descriptor: | 6-ethyl-5-{(3S)-3-[2-methoxy-5-(pyridin-4-yl)phenyl]but-1-yn-1-yl}pyrimidine-2,4-diamine, Dihydrofolate reductase, GLYCEROL, ... | | Authors: | Reeve, S.M, Wright, D.L. | | Deposit date: | 2018-12-13 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.243 Å) | | Cite: | Structure-Guided In Vitro to In Vivo Pharmacokinetic Optimization of Propargyl-Linked Antifolates.
Drug Metab.Dispos., 47, 2019
|
|
6DDP
 
 | | Mycobacterium tuberculosis Dihydrofolate Reductase complexed with beta-NADPH and 3'-[(2R)-4-(2,4-diamino-6-ethylpyrimidin-5-yl)but-3-yn-2-yl]-5'-methoxy[1,1'-biphenyl]-4-carboxylic acid | | Descriptor: | 3'-[(2R)-4-(2,4-diamino-6-ethylpyrimidin-5-yl)but-3-yn-2-yl]-5'-methoxy[1,1'-biphenyl]-4-carboxylic acid, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hajian, B, Wright, D. | | Deposit date: | 2018-05-10 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Drugging the Folate Pathway in Mycobacterium tuberculosis: The Role of Multi-targeting Agents.
Cell Chem Biol, 26, 2019
|
|
6DE5
 
 | |
2XH6
 
 | | Clostridium perfringens enterotoxin | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, HEAT-LABILE ENTEROTOXIN B CHAIN, octyl beta-D-glucopyranoside | | Authors: | Briggs, D.C, Naylor, C.E, Smedley III, J.G, MCClane, B.A, Basak, A.K. | | Deposit date: | 2010-06-09 | | Release date: | 2011-04-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structure of the Food-Poisoning Clostridium Perfringens Enterotoxin Reveals Similarity to the Aerolysin-Like Pore-Forming Toxins
J.Mol.Biol., 413, 2011
|
|
6DDW
 
 | | Mycobacterium tuberculosis Dihydrofolate Reductase complexed with beta-NADPH and N-(4-{[(2-amino-4-oxo-3,4-dihydropteridin-6-yl)methyl]amino}-2-hydroxybenzene-1-carbonyl)-L-glutamic acid | | Descriptor: | Dihydrofolate reductase, GLYCEROL, N-(4-{[(2-amino-4-oxo-3,4-dihydropteridin-6-yl)methyl]amino}-2-hydroxybenzene-1-carbonyl)-L-glutamic acid, ... | | Authors: | Hajian, B, Scocchera, E, Wright, D. | | Deposit date: | 2018-05-10 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Drugging the Folate Pathway in Mycobacterium tuberculosis: The Role of Multi-targeting Agents.
Cell Chem Biol, 26, 2019
|
|
1J73
 
 | | Crystal structure of an unstable insulin analog with native activity. | | Descriptor: | ZINC ION, insulin a, insulin b | | Authors: | Wan, Z, Zhao, M, Nakagawa, S, Jia, W, Weiss, M.A. | | Deposit date: | 2001-05-15 | | Release date: | 2001-05-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Non-standard insulin design: structure-activity relationships at the periphery of the insulin receptor.
J.Mol.Biol., 315, 2002
|
|
5ECC
 
 | | Klebsiella pneumoniae DfrA1 complexed with NADPH and 6-ethyl-5-(3-(2-methoxy-5-(pyridin-4-yl)phenyl)prop-1-yn-1-yl)pyrimidine-2,4-diamine | | Descriptor: | 6-ethyl-5-{3-[2-methoxy-5-(pyridin-4-yl)phenyl]prop-1-yn-1-yl}pyrimidine-2,4-diamine, CALCIUM ION, Dehydrofolate reductase type I, ... | | Authors: | Lombardo, M.N, Anderson, A.C. | | Deposit date: | 2015-10-20 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal Structures of Trimethoprim-Resistant DfrA1 Rationalize Potent Inhibition by Propargyl-Linked Antifolates.
ACS Infect Dis, 2, 2016
|
|
5ECX
 
 | | Klebsiella pneumoniae DfrA1 complexed with NADPH and 6-ethyl-5-(3-(6-(pyridin-4-yl)benzo[d][1,3]dioxol-4-yl)but-1-yn-1-yl)pyrimidine-2,4-diamine | | Descriptor: | 6-ethyl-5-[(3~{S})-3-(6-pyridin-4-yl-1,3-benzodioxol-4-yl)but-1-ynyl]pyrimidine-2,4-diamine, Dehydrofolate reductase type I, GLYCEROL, ... | | Authors: | Lombardo, M.N, Anderson, A.C. | | Deposit date: | 2015-10-20 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structures of Trimethoprim-Resistant DfrA1 Rationalize Potent Inhibition by Propargyl-Linked Antifolates.
ACS Infect Dis, 2, 2016
|
|
9D52
 
 | | Structure of PAK4 in complex with compound 18 | | Descriptor: | 1,2-ETHANEDIOL, 2-cyano-N-[3-({6-[(5-cyclopropyl-1,3-thiazol-2-yl)amino]pyrazin-2-yl}amino)bicyclo[1.1.1]pentan-1-yl]acetamide, Serine/threonine-protein kinase PAK 4 | | Authors: | Boone, C, Suto, R, Olland, A. | | Deposit date: | 2024-08-13 | | Release date: | 2025-04-02 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.449 Å) | | Cite: | Identification of a p21-activated kinase 1 (PAK1) inhibitor with 10-fold selectivity against PAK2.
Bioorg.Med.Chem.Lett., 2025
|
|
9D4W
 
 | | Structure of PAK1 in complex with compound 12 | | Descriptor: | N~2~-{[(1s,4s)-4-aminocyclohexyl]methyl}-N~4~-[5-(trifluoromethyl)-1,3-thiazol-2-yl]pyrimidine-2,4-diamine, Serine/threonine-protein kinase PAK 1 | | Authors: | Dementiev, A, Suto, R.K, Olland, A.M. | | Deposit date: | 2024-08-13 | | Release date: | 2025-04-02 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.218 Å) | | Cite: | Identification of a p21-activated kinase 1 (PAK1) inhibitor with 10-fold selectivity against PAK2.
Bioorg.Med.Chem.Lett., 2025
|
|
