8HO5
 
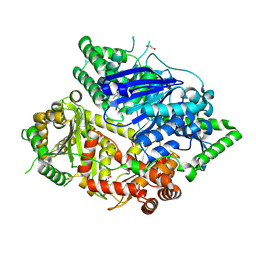 | | Falcilysin in complex with MMV665806 | | Descriptor: | (2S)-1-(2,3-dimethyl-1H-indol-1-yl)-3-{[(1S,2S)-2-methylcyclohexyl]amino}propan-2-ol, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lin, J.Q, Chung, Z, Lescar, J. | | Deposit date: | 2022-12-09 | | Release date: | 2023-12-20 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Identification of an inhibitory pocket in falcilysin provides a new avenue for malaria drug development.
Cell Chem Biol, 31, 2024
|
|
4UWT
 
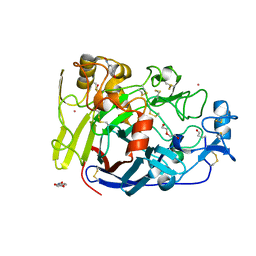 | | Hypocrea jecorina Cel7A E212Q mutant in complex with p-nitrophenyl cellobioside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLOBIOHYDROLASE CEL7A, COBALT (II) ION, ... | | Authors: | Nutt, A, Momeni, M.H, Johansson, G, Stahlberg, J. | | Deposit date: | 2014-08-14 | | Release date: | 2015-08-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Enzyme kinetics by GH7 cellobiohydrolases on chromogenic substrates is dictated by non-productive binding: insights from crystal structures and MD simulation.
Febs J., 2022
|
|
2VD7
 
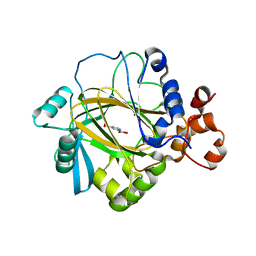 | | Crystal Structure of JMJD2A complexed with inhibitor Pyridine-2,4- dicarboxylic acid | | Descriptor: | JMJC DOMAIN-CONTAINING HISTONE DEMETHYLATION PROTEIN 3A, NICKEL (II) ION, PYRIDINE-2,4-DICARBOXYLIC ACID, ... | | Authors: | Ng, S.S, von Delft, F, Pilka, E.S, Kavanagh, K.L, McDonough, M.A, Savitsky, P, Arrowsmith, C.H, Weigelt, J, Edwards, A, Sundstrom, M, Schofield, C.J, Oppermann, U. | | Deposit date: | 2007-10-01 | | Release date: | 2007-11-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Inhibitor scaffolds for 2-oxoglutarate-dependent histone lysine demethylases.
J. Med. Chem., 51, 2008
|
|
2VNM
 
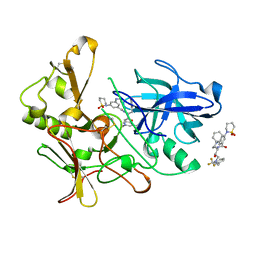 | | Human BACE-1 in complex with 3-(1,1-dioxidotetrahydro-2H-1,2-thiazin- 2-yl)-5-(ethylamino)-N-((1S,2R)-2-hydroxy-1-(phenylmethyl)-3-(((3-(trifluoromethyl)phenyl)methyl)amino)propyl)benzamide | | Descriptor: | BETA-SECRETASE 1, N-[(1S,2R)-1-benzyl-2-hydroxy-3-{[3-(trifluoromethyl)benzyl]amino}propyl]-3-(1,1-dioxido-1,2-thiazinan-2-yl)-5-(ethylamino)benzamide | | Authors: | Charrier, N, Clarke, B, Cutler, L, Demont, E, Dingwall, C, Dunsdon, R, East, P, Hawkins, J, Howes, C, Hussain, I, Jeffrey, P, Maile, G, Matico, R, Mosley, J, Naylor, A, OBrien, A, Redshaw, S, Rowland, P, Soleil, V, Smith, K.J, Sweitzer, S, Theobald, P, Vesey, D, Walter, D.S, Wayne, G. | | Deposit date: | 2008-02-05 | | Release date: | 2008-05-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Second Generation of Hydroxyethylamine Bace-1 Inhibitors: Optimizing Potency and Oral Bioavailability.
J.Med.Chem., 51, 2008
|
|
9L1S
 
 | | Structure of the HER2 (S310F) - pertuzumab (T30S/D31A) complex | | Descriptor: | Pertuzumab Fab heavy chain, Pertuzumab Fab light chain, Receptor tyrosine-protein kinase erbB-2 | | Authors: | Xu, L, Guo, J. | | Deposit date: | 2024-12-15 | | Release date: | 2025-01-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Computational-Aided Rational Mutation Design of Pertuzumab to Overcome Active HER2 Mutation S310F through Antibody-Drug Conjugates
To Be Published
|
|
3SBL
 
 | | Crystal Structure of New Delhi Metal-beta-lactamase-1 from Klebsiella pneumoniae | | Descriptor: | Beta-lactamase NDM-1, CITRIC ACID | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, J, Binkowski, T.A, Mire, J, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2011-06-05 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure of Apo- and Monometalated Forms of NDM-1 A Highly Potent Carbapenem-Hydrolyzing Metallo-beta-Lactamase
Plos One, 6, 2011
|
|
4CY3
 
 | | Crystal structure of the NSL1-WDS complex. | | Descriptor: | CG4699, ISOFORM D, GLYCEROL, ... | | Authors: | Dias, J, Brettschneider, J, Cusack, S, Kadlec, J. | | Deposit date: | 2014-04-10 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Analysis of the Kansl1/Wdr5/Kansl2 Complex Reveals that Wdr5 is Required for Efficient Assembly and Chromatin Targeting of the Nsl Complex.
Genes Dev., 28, 2014
|
|
5DDA
 
 | | Menin in complex with MI-333 | | Descriptor: | 1,2-ETHANEDIOL, 4-{4-[5-(fluoromethyl)-1,3,4-thiadiazol-2-yl]piperazin-1-yl}-6-(2,2,2-trifluoroethyl)thieno[2,3-d]pyrimidine, DIMETHYL SULFOXIDE, ... | | Authors: | Pollock, J, Dmitry, B, Cierpicki, T, Grembecka, J. | | Deposit date: | 2015-08-24 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Rational Design of Orthogonal Multipolar Interactions with Fluorine in Protein-Ligand Complexes.
J.Med.Chem., 58, 2015
|
|
2VHU
 
 | | P4 PROTEIN FROM BACTERIOPHAGE PHI12 K241C mutant in complex with ADP and MgCl | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NTPASE P4 | | Authors: | Kainov, D.E, Mancini, E.J, Telenius, J, Lisal, J, Grimes, J.M, Bamford, D.H, Stuart, D.I, Tuma, R. | | Deposit date: | 2007-11-25 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Basis of Mechanochemical Coupling in a Hexameric Molecular Motor.
J.Biol.Chem., 283, 2008
|
|
5DDJ
 
 | | Crystal structure of recombinant foot-and-mouth-disease virus O1M-S2093Y empty capsid | | Descriptor: | Foot and mouth disease virus, VP1, VP2, ... | | Authors: | Kotecha, A, Seago, J, Scott, K, Burman, A, Loureiro, S, Ren, J, Porta, C, Ginn, H.M, Jackson, T, Perez-Martin, E, Siebert, C.A, Paul, G, Huiskonen, J.T, Jones, I.M, Esnouf, R.M, Fry, E.E, Maree, F.F, Charleston, B, Stuart, D.I. | | Deposit date: | 2015-08-25 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure-based energetics of protein interfaces guides foot-and-mouth disease virus vaccine design.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5D8A
 
 | | Crystal structure of recombinant foot-and-mouth-disease virus A22-H2093F empty capsid | | Descriptor: | VP1, VP2, VP3, ... | | Authors: | Kotecha, A, Seago, J, Scott, K, Burman, A, Loureiro, S, Ren, J, Porta, C, Ginn, H.M, Jackson, T, Perez-Martin, E, Siebert, C.A, Paul, G, Huiskonen, J.T, Jones, I.M, Esnouf, R.M, Fry, E.E, Maree, F.F, Charleston, B, Stuart, D.I. | | Deposit date: | 2015-08-16 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based energetics of protein interfaces guides foot-and-mouth disease virus vaccine design.
Nat.Struct.Mol.Biol., 22, 2015
|
|
2IAT
 
 | | Crystal structure of squid ganglion DFPase W244L mutant | | Descriptor: | CALCIUM ION, Diisopropylfluorophosphatase | | Authors: | Scharff, E.I, Koepke, J, Fritzsch, G, Luecke, C, Rueterjans, H. | | Deposit date: | 2006-09-08 | | Release date: | 2006-09-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of diisopropylfluorophosphatase from Loligo vulgaris
Structure, 9, 2001
|
|
6ETJ
 
 | | HUMAN PFKFB3 IN COMPLEX WITH KAN0438241 | | Descriptor: | 4-[[3-(5-fluoranyl-2-oxidanyl-phenyl)phenyl]sulfonylamino]-2-oxidanyl-benzoic acid, 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3, ... | | Authors: | Gustafsson, N.M.S, Lundback, T, Farnegardh, K, Groth, P, Wiitta, E, Jonsson, M, Hallberg, K, Pennisi, R, Huguet Ninou, A, Martinsson, J, Norstrom, C, Schultz, J, Andersson, M, Markova, N, Marttila, P, Norin, M, Olin, T, Helleday, T. | | Deposit date: | 2017-10-26 | | Release date: | 2018-11-07 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Targeting PFKFB3 radiosensitizes cancer cells and suppresses homologous recombination.
Nat Commun, 9, 2018
|
|
3S1Y
 
 | | AMP-C BETA-LACTAMASE (PSEUDOMONAS AERUGINOSA) in complex with a beta-lactamase inhibitor | | Descriptor: | Beta-lactamase, CHLORIDE ION, ISOPROPYL ALCOHOL, ... | | Authors: | Scapin, G, Lu, J, Fitzgerald, P.M.D, Sharma, N. | | Deposit date: | 2011-05-16 | | Release date: | 2011-06-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Side chain SAR of bicyclic Beta-lactamase inhibitors (BLIs). 2. N-Alkylated and open chain analogs of MK-8712
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3CXN
 
 | | Structure of the Urease Accessory Protein UreF from Helicobacter pylori | | Descriptor: | GLYCEROL, Urease accessory protein ureF | | Authors: | Lam, R, Johns, K, Romanov, V, Dong, A, Wu-Brown, J, Guthrie, J, Dharamsi, A, Thambipillai, D, Mansoury, K, Edwards, A.M, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2008-04-24 | | Release date: | 2009-05-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of a truncated urease accessory protein UreF from Helicobacter pylori.
Proteins, 78, 2010
|
|
2VD6
 
 | | Human adenylosuccinate lyase in complex with its substrate N6-(1,2- Dicarboxyethyl)-AMP, and its products AMP and fumarate. | | Descriptor: | 2-[9-(3,4-DIHYDROXY-5-PHOSPHONOOXYMETHYL-TETRAHYDRO-FURAN-2-YL)-9H-PURIN-6-YLAMINO]-SUCCINIC ACID, ADENOSINE MONOPHOSPHATE, ADENYLOSUCCINATE LYASE, ... | | Authors: | Stenmark, P, Moche, M, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Dahlgren, L.G, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg-schiavone, L, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Nilsson, M, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Sundstrom, M, Thorsell, A.G, Tresaugues, L, van den Berg, S, Weigelt, J, Welin, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-09-30 | | Release date: | 2007-10-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human Adenylosuccinate Lyase in Complex with its Substrate N6-(1,2-Dicarboxyethyl)-AMP, and its Products AMP and Fumarate.
To be Published
|
|
4UAG
 
 | | UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE | | Descriptor: | SULFATE ION, UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE, UNKNOWN ATOM OR ION, ... | | Authors: | Bertrand, J, Auger, G, Martin, L, Fanchon, E, Blanot, D, Le Beller, D, Van Heijenoort, J, Dideberg, O. | | Deposit date: | 1999-03-09 | | Release date: | 2000-03-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Determination of the MurD mechanism through crystallographic analysis of enzyme complexes.
J.Mol.Biol., 289, 1999
|
|
6F22
 
 | | Complex between MTH1 and compound 29 (a 4-amino-2,7-diazaindole derivative) | | Descriptor: | (3~{S})-3-phenyl-4-(2~{H}-pyrazolo[3,4-b]pyridin-4-yl)morpholine, 7,8-dihydro-8-oxoguanine triphosphatase, SULFATE ION | | Authors: | Viklund, J, Talagas, A, Tresaugues, L, Andersson, M, Ericsson, U, Forsblom, R, Ginman, T, Hallberg, K, Lindstrom, J, Persson, L, Silvander, C, Rahm, F. | | Deposit date: | 2017-11-23 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Creation of a Novel Class of Potent and Selective MutT Homologue 1 (MTH1) Inhibitors Using Fragment-Based Screening and Structure-Based Drug Design.
J. Med. Chem., 61, 2018
|
|
1TVU
 
 | |
5XXF
 
 | | Crystal structure of Poz1, Tpz1 and Rap1 | | Descriptor: | Protection of telomeres protein poz1, Protection of telomeres protein tpz1, Rap1, ... | | Authors: | Xue, J, Chen, H, Wu, J, Lei, M. | | Deposit date: | 2017-07-03 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the fission yeast S. pombe telomeric Tpz1-Poz1-Rap1 complex.
Cell Res., 27, 2017
|
|
3JZH
 
 | | EED-H3K79me3 | | Descriptor: | HISTONE PEPTIDE, Polycomb protein EED | | Authors: | Xu, C, Bian, C.B, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-23 | | Release date: | 2009-12-15 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Binding of different histone marks differentially regulates the activity and specificity of polycomb repressive complex 2 (PRC2).
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5DDF
 
 | | Menin in complex with MI-273 | | Descriptor: | 4-[4-(5,5-dimethyl-4,5-dihydro-1,3-thiazol-2-yl)piperazin-1-yl]-6-(pentafluoroethyl)thieno[2,3-d]pyrimidine, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Pollock, J, Dmitry, B, Cierpicki, T, Grembecka, J. | | Deposit date: | 2015-08-24 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Rational Design of Orthogonal Multipolar Interactions with Fluorine in Protein-Ligand Complexes.
J.Med.Chem., 58, 2015
|
|
5DDS
 
 | | Crystal structure of aminotransferase CrmG from Actinoalloteichus sp. WH1-2216-6 in complex with PLP | | Descriptor: | ACETIC ACID, CrmG, GLYCEROL, ... | | Authors: | Xu, J, Feng, Z, Liu, J. | | Deposit date: | 2015-08-25 | | Release date: | 2016-08-10 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Biochemical and Structural Insights into the Aminotransferase CrmG in Caerulomycin Biosynthesis
Acs Chem.Biol., 11, 2016
|
|
4CY5
 
 | | Crystal structure of the NSL1-WDS-NSL2 complex. | | Descriptor: | CG4699, ISOFORM D, DIM GAMMA-TUBULIN 1, ... | | Authors: | Dias, J, Brettschneider, J, Cusack, S, Kadlec, J. | | Deposit date: | 2014-04-10 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Analysis of the Kansl1/Wdr5/Kansl2 Complex Reveals that Wdr5 is Required for Efficient Assembly and Chromatin Targeting of the Nsl Complex.
Genes Dev., 28, 2014
|
|
2IWA
 
 | | Unbound glutaminyl cyclotransferase from Carica papaya. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLUTAMINE CYCLOTRANSFERASE, ... | | Authors: | Guevara, T, Mallorqui-Fernandez, N, Garcia-Castellanos, R, Petersen, G.E, Lauritzen, C, Pedersen, J, Arnau, J, Gomis-Ruth, F.X, Sola, M. | | Deposit date: | 2006-06-27 | | Release date: | 2006-07-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Papaya Glutamine Cyclotransferase Shows a Singular Five-Fold Beta-Propeller Architecture that Suggests a Novel Reaction Mechanism.
Biol.Chem., 387, 2006
|
|
