8IF5
 
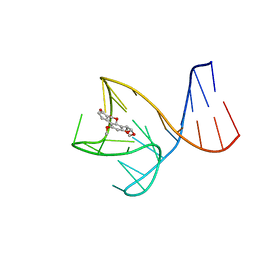 | | AFB1-AF26 APTAMER COMPLEX | | Descriptor: | AFB1 DNA aptamer (26-MER), AFLATOXIN B1 | | Authors: | Xu, G.H, Wang, C, Li, C.G. | | Deposit date: | 2023-02-17 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for high-affinity recognition of aflatoxin B1 by a DNA aptamer.
Nucleic Acids Res., 51, 2023
|
|
6K3K
 
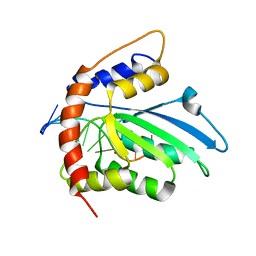 | | Solution structure of APOBEC3G-CD2 with ssDNA, Product B | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, DNA/RNA (5'-D(*AP*TP*TP*CP*UP*(ICY)P*AP*AP*TP*T)-3'), ZINC ION | | Authors: | Cao, C, Yan, X, Lan, W, Wang, C. | | Deposit date: | 2019-05-19 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Investigations on the Interactions between Cytidine Deaminase Human APOBEC3G and DNA.
Chem Asian J, 14, 2019
|
|
6K3J
 
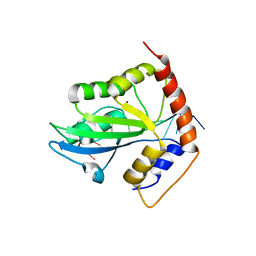 | | Solution structure of APOBEC3G-CD2 with ssDNA, Product A | | Descriptor: | DNA (5'-D(*AP*TP*TP*CP*UP*(IUR)P*AP*AP*TP*T)-3'), DNA dC->dU-editing enzyme APOBEC-3G, ZINC ION | | Authors: | Cao, C, Yan, X, Lan, W, Wang, C. | | Deposit date: | 2019-05-19 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Investigations on the Interactions between Cytidine Deaminase Human APOBEC3G and DNA.
Chem Asian J, 14, 2019
|
|
2MOX
 
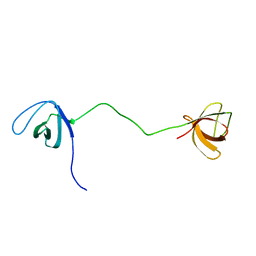 | | solution structure of tandem SH3 domain of Sorbin and SH3 domain-containing protein 1 | | Descriptor: | Sorbin and SH3 domain-containing protein 1 | | Authors: | Zhao, D, Wang, C, Zhang, J, Wu, J, Shi, Y, Zhang, Z, Gong, Q. | | Deposit date: | 2014-05-07 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural investigation of the interaction between the tandem SH3 domains of c-Cbl-associated protein and vinculin
J.Struct.Biol., 187, 2014
|
|
7BYJ
 
 | | Crystal structure of the FERM domain of FRMPD4 | | Descriptor: | FERM and PDZ domain-containing protein 4 | | Authors: | Lin, L, Wang, M, Wang, C, Zhu, J. | | Deposit date: | 2020-04-23 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure of the FERM domain of a neural scaffold protein FRMPD4 implicated in X-linked intellectual disability.
Biochem.J., 477, 2020
|
|
7XH6
 
 | | Crystal structure of CBP bromodomain liganded with CCS1477 | | Descriptor: | (6S)-1-[3,4-bis(fluoranyl)phenyl]-6-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-(4-methoxycyclohexyl)benzimidazol-2-yl]piperidin-2-one, CREB-binding protein, DIMETHYL SULFOXIDE, ... | | Authors: | Xu, H, Xiang, Q, Wang, C, Zhang, C, Luo, G, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-04-07 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights revealed by the cocrystal structure of CCS1477 in complex with CBP bromodomain
Biochem.Biophys.Res.Commun., 623, 2022
|
|
7XHE
 
 | | Crystal structure of CBP bromodomain liganded with CCS151 | | Descriptor: | (6S)-6-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(3R)-1-methylsulfonylpyrrolidin-3-yl]benzimidazol-2-yl]-1-(3-fluoranyl-4-methoxy-phenyl)piperidin-2-one, 1,2-ETHANEDIOL, CREB-binding protein | | Authors: | Xu, H, Xiang, Q, Wang, C, Zhang, C, Luo, G, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-04-08 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural insights revealed by the cocrystal structure of CCS1477 in complex with CBP bromodomain
Biochem.Biophys.Res.Commun., 623, 2022
|
|
7XI0
 
 | | Crystal structure of CBP bromodomain liganded with CCS150 | | Descriptor: | (6S)-1-(3-chloranyl-4-methoxy-phenyl)-6-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(3R)-1-methylsulfonylpyrrolidin-3-yl]benzimidazol-2-yl]piperidin-2-one, CREB-binding protein, GLYCEROL | | Authors: | Xu, H, Xiang, Q, Wang, C, Zhang, C, Luo, G, Wu, X, Zhang, Y, Xu, Y. | | Deposit date: | 2022-04-11 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural insights revealed by the cocrystal structure of CCS1477 in complex with CBP bromodomain
Biochem.Biophys.Res.Commun., 623, 2022
|
|
7V1U
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor ZJ12 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(3-ethyl-6-methoxy-1,2-benzoxazol-5-yl)-2-methoxy-benzenesulfonamide | | Authors: | Zhang, M, Wang, C, Zhang, C, Zhang, Y, Xu, Y. | | Deposit date: | 2021-08-06 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Design, synthesis and pharmacological characterization of N-(3-ethylbenzo[d]isoxazol-5-yl) sulfonamide derivatives as BRD4 inhibitors against acute myeloid leukemia.
Acta Pharmacol.Sin., 43, 2022
|
|
7V2J
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor 33 | | Descriptor: | Bromodomain-containing protein 4, ~{N}-(3-ethyl-6-methoxy-1,2-benzoxazol-5-yl)-4-methoxy-benzenesulfonamide | | Authors: | Zhang, M, Wang, C, Zhang, C, Zhang, Y, Xu, Y. | | Deposit date: | 2021-08-09 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Design, synthesis and pharmacological characterization of N-(3-ethylbenzo[d]isoxazol-5-yl) sulfonamide derivatives as BRD4 inhibitors against acute myeloid leukemia.
Acta Pharmacol.Sin., 43, 2022
|
|
5Y4D
 
 | | Crystal Structure of AnkB Ankyrin Repeats in Complex with AnkR/AnkB Chimeric Autoinhibition Segment | | Descriptor: | Ankyrin-1,Ankyrin-2,Ankyrin-2, SULFATE ION | | Authors: | Chen, K, Li, J, Wang, C, Wei, Z, Zhang, M. | | Deposit date: | 2017-08-03 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Autoinhibition of ankyrin-B/G membrane target bindings by intrinsically disordered segments from the tail regions.
Elife, 6, 2017
|
|
5Y4F
 
 | | Crystal Structure of AnkB Ankyrin Repeats R13-24 in complex with autoinhibition segment AI-c | | Descriptor: | ACETATE ION, Ankyrin-2, CALCIUM ION | | Authors: | Chen, K, Li, J, Wang, C, Wei, Z, Zhang, M. | | Deposit date: | 2017-08-03 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Autoinhibition of ankyrin-B/G membrane target bindings by intrinsically disordered segments from the tail regions.
Elife, 6, 2017
|
|
7CDX
 
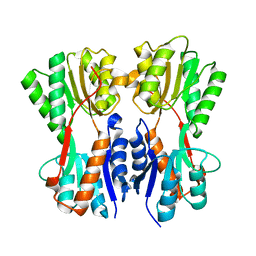 | | Complex STRUCTURE OF A NOVEL VIRULENCE REGULATION FACTOR SghR with its effector sucrose | | Descriptor: | LacI-type transcription factor, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Ye, F.Z, Wang, C, Yan, X.F, Zhang, L.H, Gao, Y.G. | | Deposit date: | 2020-06-20 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Structural basis of a novel repressor, SghR, controllingAgrobacteriuminfection by cross-talking to plants.
J.Biol.Chem., 295, 2020
|
|
5YCO
 
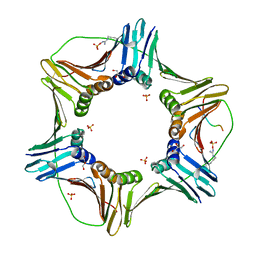 | | Complex structure of PCNA with UHRF2 | | Descriptor: | E3 ubiquitin-protein ligase UHRF2, GLYCEROL, Proliferating cell nuclear antigen, ... | | Authors: | Wu, M, Chen, W, Hang, T, Wang, C, Zhang, X, Zang, J. | | Deposit date: | 2017-09-07 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Structure insights into the molecular mechanism of the interaction between UHRF2 and PCNA.
Biochem. Biophys. Res. Commun., 494, 2017
|
|
5Z6Q
 
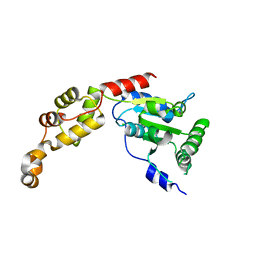 | | Crystal structure of AAA of Spastin | | Descriptor: | CHLORIDE ION, Spastin | | Authors: | Lin, Z, Wang, C, Shen, Y. | | Deposit date: | 2018-01-25 | | Release date: | 2018-12-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The AAA protein spastin possesses two levels of basal ATPase activity
FEBS Lett., 592, 2018
|
|
5Z6R
 
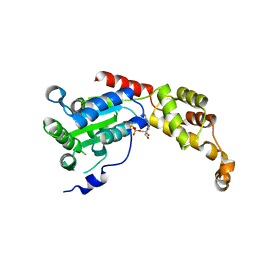 | | SPASTIN AAA WITH ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Spastin | | Authors: | Lin, Z, Wang, C, Shen, Y. | | Deposit date: | 2018-01-25 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Aaa Family Protein Spastin Severs A Microtubule
To Be Published
|
|
6AJK
 
 | | Crystal structure of TFB1M and h45 in homo sapiens | | Descriptor: | Dimethyladenosine transferase 1, mitochondrial, RNA (28-MER) | | Authors: | Liu, X, Shen, S, Wu, P, Li, F, Wang, C, Gong, Q, Wu, J, Zhang, H, Shi, Y. | | Deposit date: | 2018-08-28 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Structural insights into dimethylation of 12S rRNA by TFB1M: indispensable role in translation of mitochondrial genes and mitochondrial function.
Nucleic Acids Res., 47, 2019
|
|
6AO4
 
 | |
6AO3
 
 | |
7CE1
 
 | | Complex STRUCTURE OF TRANSCRIPTION FACTOR SghR with its COGNATE DNA | | Descriptor: | LacI-type transcription factor, promoter DNA | | Authors: | Ye, F.Z, Wang, C, Yan, X.F, Zhang, L.H, Gao, Y.G. | | Deposit date: | 2020-06-21 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of a novel repressor, SghR, controllingAgrobacteriuminfection by cross-talking to plants.
J.Biol.Chem., 295, 2020
|
|
7ECA
 
 | |
7XIJ
 
 | | Crystal structure of CBP bromodomain liganded with Y08175 | | Descriptor: | 3-[(1-ethanoyl-5-methoxy-indol-3-yl)carbonylamino]-4-fluoranyl-5-(1-methylpyrazol-4-yl)benzoic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Xiang, Q, Wang, C, Wu, T, Zhang, Y, Zhang, C, Luo, G, Wu, X, Shen, H, Xu, Y. | | Deposit date: | 2022-04-13 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of CBP bromodomain liganded with Y08175
To Be Published
|
|
7XNE
 
 | | Crystal structure of CBP bromodomain liganded with Y08284 | | Descriptor: | CREB-binding protein, GLYCEROL, N-[3-(1-cyclopropylpyrazol-4-yl)-2-fluoranyl-5-[(1S)-1-oxidanylethyl]phenyl]-3-ethanoyl-7-methoxy-indolizine-1-carboxamide | | Authors: | Xiang, Q, Wang, C, Wu, T, Zhang, C, Hu, Q, Luo, G, Hu, J, Zhuang, X, Zou, L, Shen, H, Wu, X, Zhang, Y, Kong, X, Xu, Y. | | Deposit date: | 2022-04-28 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 1-(Indolizin-3-yl)ethan-1-ones as CBP Bromodomain Inhibitors for the Treatment of Prostate Cancer.
J.Med.Chem., 65, 2022
|
|
7CDV
 
 | | STRUCTURE OF A NOVEL VIRULENCE REGULATION FACTOR SghR | | Descriptor: | LacI-type transcription factor | | Authors: | Ye, F.Z, Wang, C, Yan, X.F, Zhang, L.H, Gao, Y.G. | | Deposit date: | 2020-06-20 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of a novel repressor, SghR, controllingAgrobacteriuminfection by cross-talking to plants.
J.Biol.Chem., 295, 2020
|
|
2LZ3
 
 | |
