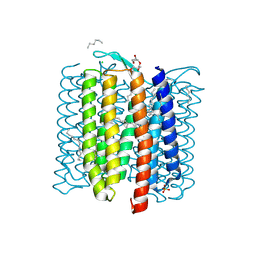
8H79
 
 | |
5WZP
 
 | | Alpha-N-acetylgalactosaminidase NagBb from Bifidobacterium bifidum - ligand free | | Descriptor: | Alpha-N-acetylgalactosaminidase, CALCIUM ION, ZINC ION | | Authors: | Sato, M, Arakawa, T, Ashida, H, Fushinobu, S. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | The first crystal structure of a family 129 glycoside hydrolase from a probiotic bacterium reveals critical residues and metal cofactors
J. Biol. Chem., 292, 2017
|
|
7NAZ
 
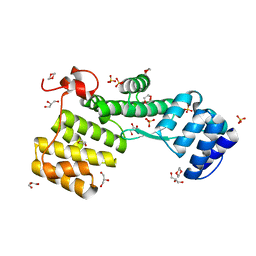 | | TPR-rich domain of EccA3 from M. smegmatis | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, ESX-3 secretion system protein EccA3, GLYCEROL, ... | | Authors: | Crosskey, T.D, Wilmanns, M. | | Deposit date: | 2021-01-25 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of TPR-rich domain of M. smegmatis EccA3
To Be Published
|
|
5WRC
 
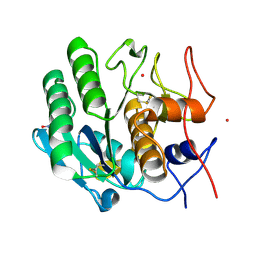 | | Crystal structure of proteinase K from Engyodontium album | | Descriptor: | NITRATE ION, PRASEODYMIUM ION, Proteinase K | | Authors: | Sugahara, M, Nakane, T, Suzuki, M, Masuda, T, Inoue, S, Numata, K. | | Deposit date: | 2016-12-01 | | Release date: | 2017-11-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Hydroxyethyl cellulose matrix applied to serial crystallography
Sci Rep, 7, 2017
|
|
7NKW
 
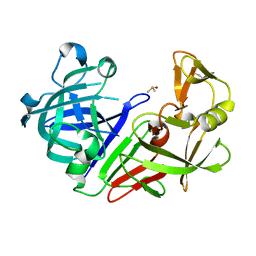 | | Endothiapepsin structure obtained at 298K after a soaking with fragment JFD03909 from a dataset collected with JUNGFRAU detector | | Descriptor: | DIMETHYL SULFOXIDE, Endothiapepsin | | Authors: | Engilberge, S, Huang, C.-Y, Leonarski, F, Wojdyla, J.A, Marsh, M, Olieric, V, Wang, M. | | Deposit date: | 2021-02-19 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Endothiapepsin structure obtained at 298K after a soaking with fragment JFD03909 from a dataset collected with JUNGFRAU detector
To Be Published
|
|
8PFL
 
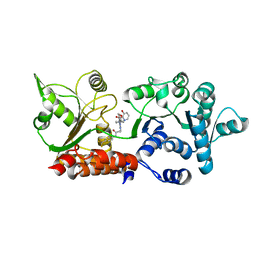 | | Crystal structure of WRN helicase domain in complex with 3 | | Descriptor: | 2-[2-(3,6-dihydro-2~{H}-pyran-4-yl)-5-ethyl-7-oxidanylidene-6-[4-(3-oxidanylpyridin-2-yl)carbonylpiperazin-1-yl]-[1,2,4]triazolo[1,5-a]pyrimidin-4-yl]-~{N}-[2-methyl-4-(trifluoromethyl)phenyl]ethanamide, Bifunctional 3'-5' exonuclease/ATP-dependent helicase WRN, ZINC ION | | Authors: | Scheufler, C, Meyer, M, Moebitz, H. | | Deposit date: | 2023-06-16 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of WRN inhibitor HRO761 with synthetic lethality in MSI cancers.
Nature, 629, 2024
|
|
5JGD
 
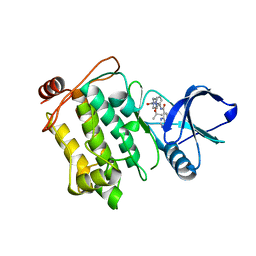 | | Crystal structure of human TAK1/TAB1 fusion protein in complex with ligand 12 | | Descriptor: | N-(2-isopropoxy-3-(4-methylpiperazine-1-carbonyl)phenyl)-4-oxo-3,4-dihydrothieno[3,2-d]pyrimidine-7-carboxamide, TAK1 kinase - TAB1 chimera fusion protein | | Authors: | Irie, M, Nakamura, M, Fukami, T.A, Matsuura, T, Morishima, K. | | Deposit date: | 2016-04-20 | | Release date: | 2016-07-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | Discovery of a potent and highly selective transforming growth factor beta receptor-associated kinase 1 (TAK1) inhibitor by structure based drug design (SBDD)
Bioorg.Med.Chem., 24, 2016
|
|
8PFO
 
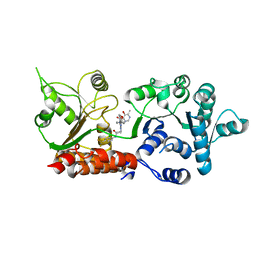 | | Crystal structure of WRN helicase domain in complex with HRO761 | | Descriptor: | Bifunctional 3'-5' exonuclease/ATP-dependent helicase WRN, ZINC ION, ~{N}-[2-chloranyl-4-(trifluoromethyl)phenyl]-2-[2-(3,6-dihydro-2~{H}-pyran-4-yl)-5-ethyl-6-[4-(6-methyl-5-oxidanyl-pyrimidin-4-yl)carbonylpiperazin-1-yl]-7-oxidanylidene-[1,2,4]triazolo[1,5-a]pyrimidin-4-yl]ethanamide | | Authors: | Scheufler, C, Meyer, M, Moebitz, H. | | Deposit date: | 2023-06-16 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of WRN inhibitor HRO761 with synthetic lethality in MSI cancers.
Nature, 629, 2024
|
|
7PVO
 
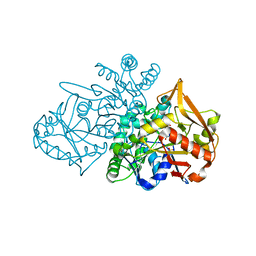 | | Adenylosuccinate Synthetase from H. pylori in complex with IMP | | Descriptor: | Adenylosuccinate synthetase, INOSINIC ACID, SULFATE ION | | Authors: | Narczyk, M, Bzowska, A, Maksymiuk, W. | | Deposit date: | 2021-10-05 | | Release date: | 2022-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The pursuit of new alternative ways to eradicate Helicobacter pylori continues: Detailed characterization of interactions in the adenylosuccinate synthetase active site.
Int.J.Biol.Macromol., 226, 2023
|
|
4UND
 
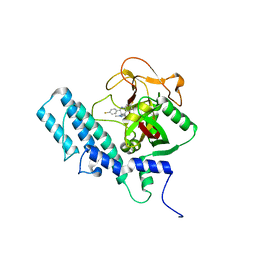 | | HUMAN ARTD1 (PARP1) - CATALYTIC DOMAIN IN COMPLEX WITH INHIBITOR TALAZOPARIB | | Descriptor: | (8S,9R)-5-fluoro-8-(4-fluorophenyl)-9-(1-methyl-1H-1,2,4-triazol-5-yl)-2,7,8,9-tetrahydro-3H-pyrido[4,3,2-de]phthalazin-3-one, POLY [ADP-RIBOSE] POLYMERASE 1, SODIUM ION | | Authors: | Karlberg, T, Thorsell, A.G, Ekblad, T, Klepsch, M, Pinto, A.F, Tresaugues, L, Moche, M, Schuler, H. | | Deposit date: | 2014-05-27 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Potency and Promiscuity in Poly(ADP-ribose) Polymerase (PARP) and Tankyrase Inhibitors.
J. Med. Chem., 60, 2017
|
|
8Q7S
 
 | | Crystal structure of the SARS-CoV-2 RBD (Wuhan) with neutralizing VHHs Ma6F06 and Re21H01 | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, GLYCEROL, ... | | Authors: | Guttler, T, Aksu, M, Gorlich, D. | | Deposit date: | 2023-08-16 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Nanobodies to multiple spike variants and inhalation of nanobody-containing aerosols neutralize SARS-CoV-2 in cell culture and hamsters.
Antiviral Res., 221, 2023
|
|
5WR8
 
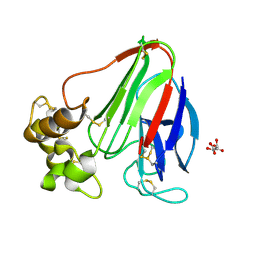 | | Thaumatin structure determined by SACLA at 1.55 Angstrom | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Masuda, T, Suzuki, M, Inoue, S, Sugahara, M. | | Deposit date: | 2016-12-01 | | Release date: | 2017-11-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Hydroxyethyl cellulose matrix applied to serial crystallography
Sci Rep, 7, 2017
|
|
6Y3C
 
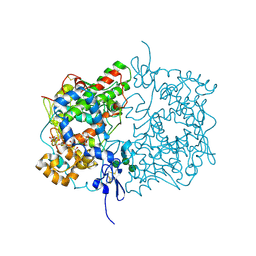 | | Human COX-1 Crystal Structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRATE ANION, Prostaglandin G/H synthase 1, ... | | Authors: | Miciaccia, M, Belviso, B.D, Iaselli, M, Ferorelli, S, Perrone, M.G, Caliandro, R, Scilimati, A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.361 Å) | | Cite: | Three-dimensional structure of human cyclooxygenase (hCOX)-1.
Sci Rep, 11, 2021
|
|
8QMH
 
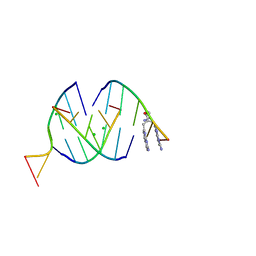 | | Crystal structure of RNA G2C4 repeats in complex with small synthetic molecule ANP77 | | Descriptor: | 3-(7-azanyl-1,8-naphthyridin-2-yl)-2-[(7-azanyl-1,8-naphthyridin-2-yl)methyl]-~{N}-(3-azanylpropyl)propanamide, CHLORIDE ION, RNA (5'-R(*GP*GP*CP*CP*CP*C)-3') | | Authors: | Kiliszek, A, Ryczek, M, Blaszczyk, L. | | Deposit date: | 2023-09-22 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Antisense RNA C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms a triplex-like structure and binds small synthetic ligand.
Nucleic Acids Res., 2024
|
|
6YOE
 
 | | Structure of Lysozyme from SiN IMISX setup collected by still serial crystallography on crystals prelocated by 2D X-ray phase-contrast imaging | | Descriptor: | 2-(2-ETHOXYETHOXY)ETHANOL, ACETIC ACID, BROMIDE ION, ... | | Authors: | Huang, C.-Y, Martiel, I, Villanueva-Perez, P, Panepucci, E, Caffrey, M, Wang, M. | | Deposit date: | 2020-04-14 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Low-dose in situ prelocation of protein microcrystals by 2D X-ray phase-contrast imaging for serial crystallography.
Iucrj, 7, 2020
|
|
7QDH
 
 | | SARS-CoV-2 S protein S:D614G mutant 1-up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Ginex, T, Marco-Marin, C, Wieczor, M, Mata, C.P, Krieger, J, Lopez-Redondo, M.L, Frances-Gomez, C, Ruiz-Rodriguez, P, Melero, R, Sanchez-Sorzano, C.O, Martinez, M, Gougeard, N, Forcada-Nadal, A, Zamora-Caballero, S, Gozalbo-Rovira, R, Sanz-Frasquet, C, Bravo, J, Rubio, V, Marina, A, Geller, R, Comas, I, Gil, C, Coscolla, M, Orozco, M, LLacer, J.L, Carazo, J.M. | | Deposit date: | 2021-11-27 | | Release date: | 2022-05-25 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The structural role of SARS-CoV-2 genetic background in the emergence and success of spike mutations: The case of the spike A222V mutation.
Plos Pathog., 18, 2022
|
|
7QDG
 
 | | SARS-CoV-2 S protein S:A222V + S:D614G mutant 1-up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ginex, T, Marco-Marin, C, Wieczor, M, Mata, C.P, Krieger, J, Lopez-Redondo, M.L, Frances-Gomez, C, Ruiz-Rodriguez, P, Melero, R, Sanchez-Sorzano, C.O, Martinez, M, Gougeard, N, Forcada-Nadal, A, Zamora-Caballero, S, Gozalbo-Rovira, R, Sanz-Frasquet, C, Bravo, J, Rubio, V, Marina, A, Geller, R, Comas, I, Gil, C, Coscolla, M, Orozco, M, LLacer, J.L, Carazo, J.M. | | Deposit date: | 2021-11-27 | | Release date: | 2022-05-25 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The structural role of SARS-CoV-2 genetic background in the emergence and success of spike mutations: The case of the spike A222V mutation.
Plos Pathog., 18, 2022
|
|
7QDN
 
 | |
7QCO
 
 | | The structure of Photosystem I tetramer from Chroococcidiopsis TS-821, a thermophilic, unicellular, non-heterocyst-forming cyanobacterium | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, BETA-CAROTENE, CHLOROPHYLL A, ... | | Authors: | Semchonok, D.A, Mondal, J, Cooper, J.C, Schlum, K, Li, M, Amin, M, Sorzano, C.O.S, Ramirez-Aportela, E, Kastritis, P.L, Boekema, E.J, Guskov, A, Bruce, B.D. | | Deposit date: | 2021-11-24 | | Release date: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of a tetrameric photosystem I from Chroococcidiopsis TS-821, a thermophilic, unicellular, non-heterocyst-forming cyanobacterium.
Plant Commun., 3, 2022
|
|
7QJU
 
 | | EED in complex with PRC2 allosteric inhibitor compound 7 | | Descriptor: | CHLORIDE ION, Histone-lysine N-methyltransferase EZH2, N-(2,3-dihydro-1-benzofuran-4-ylmethyl)-8-[4-[(dimethylamino)methyl]phenyl]-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, ... | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H, Scheufler, C. | | Deposit date: | 2021-12-17 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of the Clinical Candidate MAK683: An EED-Directed, Allosteric, and Selective PRC2 Inhibitor for the Treatment of Advanced Malignancies.
J.Med.Chem., 65, 2022
|
|
7QJG
 
 | | EED in complex with PRC2 allosteric inhibitor compound 6 | | Descriptor: | CHLORIDE ION, Histone-lysine N-methyltransferase EZH2, N-(2,3-dihydro-1-benzofuran-7-ylmethyl)-8-[4-[(dimethylamino)methyl]phenyl]-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, ... | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H, Scheufler, C. | | Deposit date: | 2021-12-16 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of the Clinical Candidate MAK683: An EED-Directed, Allosteric, and Selective PRC2 Inhibitor for the Treatment of Advanced Malignancies.
J.Med.Chem., 65, 2022
|
|
7QK4
 
 | | EED in complex with PRC2 allosteric inhibitor compound 22 (MAK683) | | Descriptor: | CHLORIDE ION, Histone-lysine N-methyltransferase EZH2, N-[(5-fluoranyl-2,3-dihydro-1-benzofuran-4-yl)methyl]-8-(2-methylpyridin-3-yl)-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, ... | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H, Scheufler, C. | | Deposit date: | 2021-12-17 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Discovery of the Clinical Candidate MAK683: An EED-Directed, Allosteric, and Selective PRC2 Inhibitor for the Treatment of Advanced Malignancies.
J.Med.Chem., 65, 2022
|
|
5X9A
 
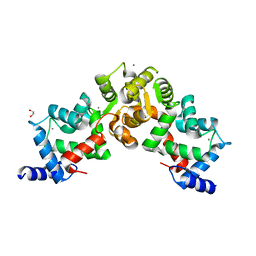 | | Crystal structure of calaxin with calcium | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Calaxin | | Authors: | Shojima, T, Hou, F, Takahashi, Y, Okai, M, Mizuno, K, Inaba, K, Miyakawa, T, Tanokura, M. | | Deposit date: | 2017-03-06 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of a Ca2+-dependent regulator of flagellar motility reveals the open-closed structural transition
Sci Rep, 8, 2018
|
|
5XFM
 
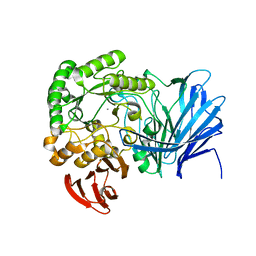 | | Crystal structure of beta-arabinopyranosidase | | Descriptor: | Alpha-glucosidase, CALCIUM ION | | Authors: | Kato, K, Okuyama, M, Yao, M. | | Deposit date: | 2017-04-10 | | Release date: | 2018-02-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A novel glycoside hydrolase family 97 enzyme: Bifunctional beta-l-arabinopyranosidase/ alpha-galactosidase from Bacteroides thetaiotaomicron.
Biochimie, 142, 2017
|
|
8FYG
 
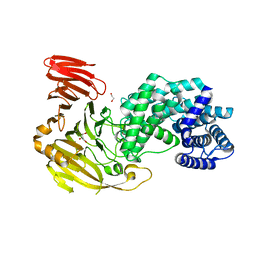 | | Crystal structure of Hyaluronate lyase A from Cutibacterium acnes | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Hyaluronate lyase | | Authors: | Katiki, M, McNally, R, Chatterjee, A, Hajam, I.A, Liu, G.Y, Murali, R. | | Deposit date: | 2023-01-26 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Functional divergence of a bacterial enzyme promotes healthy or acneic skin.
Nat Commun, 14, 2023
|
|
