8JLO
 
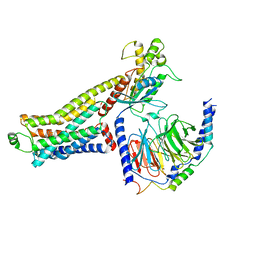 | | Ulotaront(SEP-363856)-bound hTAAR1-Gs protein complex | | Descriptor: | 1-[(7~{S})-5,7-dihydro-4~{H}-thieno[2,3-c]pyran-7-yl]-~{N}-methyl-methanamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, Z, Guo, L.L, Zhao, C, Shen, S.Y, Sun, J.P, Shao, Z.H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-15 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Ligand recognition and G-protein coupling of trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8JLR
 
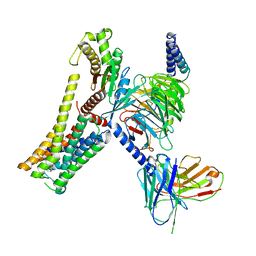 | | A77636-bound hTAAR1-Gs protein complex | | Descriptor: | (1~{S},3~{R})-3-(1-adamantyl)-1-(aminomethyl)-3,4-dihydro-1~{H}-isochromene-5,6-diol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, Z, Guo, L.L, Zhao, C, Shen, S.Y, Sun, J.P, Shao, Z.H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-15 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Ligand recognition and G-protein coupling of trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8JSP
 
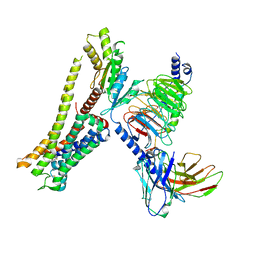 | | Ulotaront(SEP-363856)-bound Serotonin 1A (5-HT1A) receptor-Gi complex | | Descriptor: | 1-[(7~{S})-5,7-dihydro-4~{H}-thieno[2,3-c]pyran-7-yl]-~{N}-methyl-methanamine, 5-hydroxytryptamine receptor 1A, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, Z, Guo, L.L, Zhao, C, Shen, S.Y, Sun, J.P, Shao, Z.H. | | Deposit date: | 2023-06-20 | | Release date: | 2023-11-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Ligand recognition and G-protein coupling of trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8JLJ
 
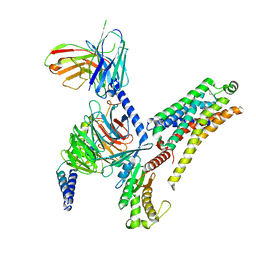 | | T1AM-bound mTAAR1-Gs protein complex | | Descriptor: | 4-[4-(2-azanylethyl)-2-iodanyl-phenoxy]phenol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, Z, Guo, L.L, Zhao, C, Shen, S.Y, Sun, J.P, Shao, Z.H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ligand recognition and G-protein coupling of trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8JSO
 
 | | AMPH-bound hTAAR1-Gs protein complex | | Descriptor: | (2S)-1-phenylpropan-2-amine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, Z, Guo, L.L, Zhao, C, Shen, S.Y, Sun, J.P, Shao, Z.H. | | Deposit date: | 2023-06-20 | | Release date: | 2023-11-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Ligand recognition and G-protein coupling of trace amine receptor TAAR1.
Nature, 624, 2023
|
|
9IJN
 
 | | STING LBD domain with an agonist DW18343 | | Descriptor: | 4-[4-(7-fluoranyl-1H-indol-3-yl)thiophen-2-yl]-4-oxidanylidene-butanoic acid, Stimulator of interferon genes protein | | Authors: | Wang, Z. | | Deposit date: | 2024-06-25 | | Release date: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Discovery of a non-nucleotide stimulator of interferon genes (STING) agonist with systemic antitumor effect.
MedComm (2020), 6, 2025
|
|
6SB1
 
 | | Crystal structure of murine perforin-2 P2 domain crystal form 1 | | Descriptor: | CHLORIDE ION, GLYCEROL, Macrophage-expressed gene 1 protein | | Authors: | Ni, T, Ginger, L, Gilbert, R.J.C. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and mechanism of bactericidal mammalian perforin-2, an ancient agent of innate immunity.
Sci Adv, 6, 2020
|
|
3VF5
 
 | | Crystal Structure of HIV-1 Protease Mutant I47V with novel P1'-Ligands GRL-02031 | | Descriptor: | (3aS,5R,6aR)-hexahydro-2H-cyclopenta[b]furan-5-yl [(1S,2R)-1-benzyl-2-hydroxy-3-([(4-methoxyphenyl)sulfonyl]{[(2R)-5-oxopyrrolidin-2-yl]methyl}amino)propyl]carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Yu, X.X, Wang, Y.F, Chang, Y.C.E, Weber, I.T. | | Deposit date: | 2012-01-09 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Potent antiviral HIV-1 protease inhibitor GRL-02031 adapts to the structures of drug resistant mutants with its P1'-pyrrolidinone ring.
J.Med.Chem., 55, 2012
|
|
3VF7
 
 | | Crystal Structure of HIV-1 Protease Mutant L76V with novel P1'-Ligands GRL-02031 | | Descriptor: | (3aS,5R,6aR)-hexahydro-2H-cyclopenta[b]furan-5-yl [(1S,2R)-1-benzyl-2-hydroxy-3-([(4-methoxyphenyl)sulfonyl]{[(2R)-5-oxopyrrolidin-2-yl]methyl}amino)propyl]carbamate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Yu, X.X, Wang, Y.F, Chang, Y.C.E, Weber, I.T. | | Deposit date: | 2012-01-09 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Potent antiviral HIV-1 protease inhibitor GRL-02031 adapts to the structures of drug resistant mutants with its P1'-pyrrolidinone ring.
J.Med.Chem., 55, 2012
|
|
3VFA
 
 | | Crystal Structure of HIV-1 Protease Mutant V82A with novel P1'-Ligands GRL-02031 | | Descriptor: | (3aS,5R,6aR)-hexahydro-2H-cyclopenta[b]furan-5-yl [(1S,2R)-1-benzyl-2-hydroxy-3-([(4-methoxyphenyl)sulfonyl]{[(2R)-5-oxopyrrolidin-2-yl]methyl}amino)propyl]carbamate, CHLORIDE ION, SODIUM ION, ... | | Authors: | Yu, X.X, Wang, Y.F, Chang, Y.C.E, Weber, I.T. | | Deposit date: | 2012-01-09 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Potent antiviral HIV-1 protease inhibitor GRL-02031 adapts to the structures of drug resistant mutants with its P1'-pyrrolidinone ring.
J.Med.Chem., 55, 2012
|
|
3VFB
 
 | | Crystal Structure of HIV-1 Protease Mutant N88D with novel P1'-Ligands GRL-02031 | | Descriptor: | (3aS,5R,6aR)-hexahydro-2H-cyclopenta[b]furan-5-yl [(1S,2R)-1-benzyl-2-hydroxy-3-([(4-methoxyphenyl)sulfonyl]{[(2R)-5-oxopyrrolidin-2-yl]methyl}amino)propyl]carbamate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Yu, X.X, Wang, Y.F, Chang, Y.C.E, Weber, I.T. | | Deposit date: | 2012-01-09 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Potent antiviral HIV-1 protease inhibitor GRL-02031 adapts to the structures of drug resistant mutants with its P1'-pyrrolidinone ring.
J.Med.Chem., 55, 2012
|
|
9O9N
 
 | | Crystal structure of PPARgamma ligand binding domain (LBD) in complex with NCoR1 corepressor peptide and inverse agonist FX-909 | | Descriptor: | (3P)-3-(5,7-difluoro-4-oxo-1,4-dihydroquinolin-2-yl)-4-(methanesulfonyl)benzonitrile, Nuclear receptor corepressor 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Laughlin, Z.T, Dong, J, Harp, J.M, Kojetin, D.J. | | Deposit date: | 2025-04-18 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of PPARgamma-mediated transcriptional repression by the covalent inverse agonist FX-909
To Be Published
|
|
9C6O
 
 | |
9DAK
 
 | |
9C45
 
 | |
9C44
 
 | |
6MS7
 
 | | Peroxisome proliferator-activated receptor gamma ligand binding domain in complex with a novel selective PPAR-gamma modulator VSP-77 | | Descriptor: | PGC1 LXXLL motif, Peroxisome proliferator-activated receptor gamma, {[(1S)-1-(4-chlorophenyl)octyl]oxy}acetic acid | | Authors: | Yi, W, Jiang, H, Zhou, X.E, Shi, J, Zhao, G, Zhang, X, Sun, Y, Suino-Powell, K, Li, J, Li, J, Melcher, K, Xu, H.E. | | Deposit date: | 2018-10-16 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Identification and structural insight of an effective PPAR gamma modulator with improved therapeutic index for anti-diabetic drug discovery.
Chem Sci, 11, 2020
|
|
3G0E
 
 | | KIT kinase domain in complex with sunitinib | | Descriptor: | Mast/stem cell growth factor receptor, N-[2-(diethylamino)ethyl]-5-[(Z)-(5-fluoro-2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]-2,4-dimethyl-1H-pyrrole-3-carbo xamide | | Authors: | Gajiwala, K.S, Wu, J.C, Lunney, E.A, Gemetri, G.D. | | Deposit date: | 2009-01-27 | | Release date: | 2009-02-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | KIT kinase mutants show unique mechanisms of drug resistance to imatinib and sunitinib in gastrointestinal stromal tumor patients.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7Q2J
 
 | | Quaternary Complex of human WDR5 and pVHL:ElonginC:ElonginB bound to PROTAC Homer | | Descriptor: | Elongin-B, Elongin-C, N-[5-[4-[[5-[[(2S)-3,3-dimethyl-1-[(2S,4R)-2-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methylcarbamoyl]-4-oxidanyl-pyrrolidin-1-yl]-1-oxidanylidene-butan-2-yl]amino]-5-oxidanylidene-pentyl]carbamoyl]phenyl]-2-(4-methylpiperazin-1-yl)phenyl]-6-oxidanylidene-4-(trifluoromethyl)-1H-pyridine-3-carboxamide, ... | | Authors: | Kraemer, A, Doelle, A, Schwalm, M.P, Adhikari, B, Wolf, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-10-25 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Tracking the PROTAC degradation pathway in living cells highlights the importance of ternary complex measurement for PROTAC optimization.
Cell Chem Biol, 30, 2023
|
|
8WC3
 
 | | Cryo-EM structure of the SEP363856-bound mTAAR1-Gs complex | | Descriptor: | 1-[(7~{S})-5,7-dihydro-4~{H}-thieno[2,3-c]pyran-7-yl]-~{N}-methyl-methanamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC4
 
 | | Cryo-EM structure of the ZH8651-bound mTAAR1-Gs complex | | Descriptor: | 2-(4-bromophenyl)ethanamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC7
 
 | | Cryo-EM structure of the ZH8667-bound mTAAR1-Gs complex | | Descriptor: | 2-[4-(3-fluorophenyl)phenyl]ethanamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC9
 
 | | Cryo-EM structure of the ZH8651-bound mTAAR1-Gq complex | | Descriptor: | 2-(4-bromophenyl)ethanamine, Engineered G-alpha-q subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC8
 
 | | Cryo-EM structure of the ZH8651-bound hTAAR1-Gs complex | | Descriptor: | 2-(4-bromophenyl)ethanamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WCA
 
 | | Cryo-EM structure of the PEA-bound hTAAR1-Gs complex | | Descriptor: | 2-PHENYLETHYLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
