8JHC
 
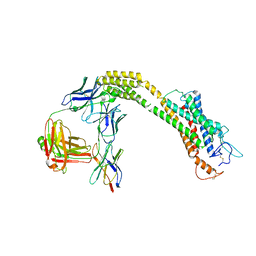 | | FZD3 in inactive state | | Descriptor: | Frizzled-3,Soluble cytochrome b562, anti-BRIL Fab Heavy chain, anti-BRIL Fab Light chain, ... | | Authors: | Xu, F, Zhang, Z. | | Deposit date: | 2023-05-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A framework for Frizzled-G protein coupling and implications to the PCP signaling pathways.
Cell Discov, 10, 2024
|
|
7UZM
 
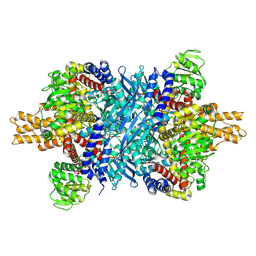 | | Glutamate dehydrogenase 1 from human liver | | Descriptor: | Glutamate dehydrogenase 1, mitochondrial | | Authors: | Zhang, Z. | | Deposit date: | 2022-05-09 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | High-resolution structural-omics of human liver enzymes.
Cell Rep, 42, 2023
|
|
1X11
 
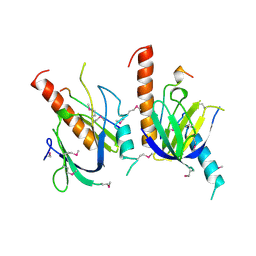 | | X11 PTB DOMAIN | | Descriptor: | 13-MER PEPTIDE, X11 | | Authors: | Lee, C.-H, Zhang, Z, Kuriyan, J. | | Deposit date: | 1997-07-28 | | Release date: | 1998-01-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Sequence-specific recognition of the internalization motif of the Alzheimer's amyloid precursor protein by the X11 PTB domain.
EMBO J., 16, 1997
|
|
6K9H
 
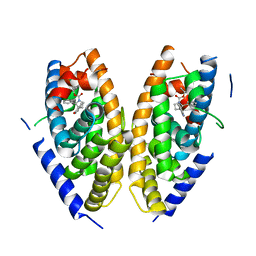 | | Human LXR-beta in complex with an agonist | | Descriptor: | Oxysterols receptor LXR-beta, ~{tert}-butyl (2'~{S},3~{S})-2-oxidanylidene-2'-phenyl-spiro[1~{H}-indole-3,3'-pyrrolidine]-1'-carboxylate | | Authors: | Zhang, Z, Zhou, H. | | Deposit date: | 2019-06-15 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of new LXR beta agonists as glioblastoma inhibitors.
Eur.J.Med.Chem., 194, 2020
|
|
6K10
 
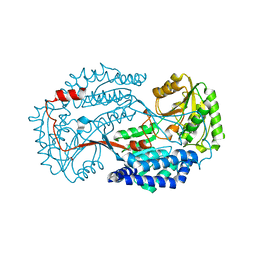 | | Non substrate bound state of Staphylococcus Aureus AldH | | Descriptor: | 1,2-ETHANEDIOL, Aldehyde dehydrogenase | | Authors: | Zhang, Z, Tao, X. | | Deposit date: | 2019-05-08 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.78962183 Å) | | Cite: | Structural Insight into the Substrate Gating Mechanism by Staphylococcus aureus Aldehyde Dehydrogenase
CCS Chemistry, 2, 2020
|
|
6K9G
 
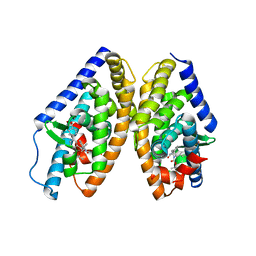 | | Human LXR-beta in complex with an agonist | | Descriptor: | Oxysterols receptor LXR-beta, ~{tert}-butyl (2'~{R},3~{R})-2'-[3-[4-(hydroxymethyl)-3-methylsulfonyl-phenyl]phenyl]-2-oxidanylidene-spiro[1~{H}-indole-3,3'-pyrrolidine]-1'-carboxylate | | Authors: | Zhang, Z, Zhou, H. | | Deposit date: | 2019-06-15 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of new LXR beta agonists as glioblastoma inhibitors.
Eur.J.Med.Chem., 194, 2020
|
|
8WX2
 
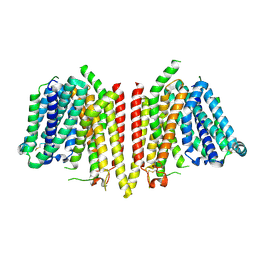 | |
8WX3
 
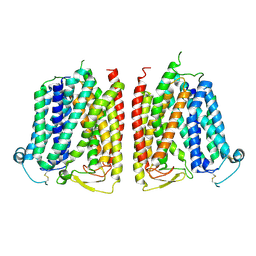 | |
8WX5
 
 | |
8WX4
 
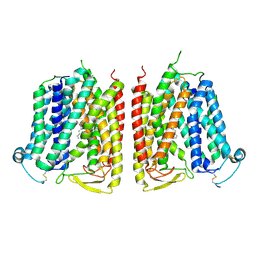 | |
6K9M
 
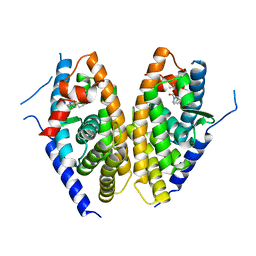 | | Human LXR-beta in complex with an agonist | | Descriptor: | Oxysterols receptor LXR-beta, ~{tert}-butyl (2'~{S},3~{S})-2-oxidanylidene-2'-propan-2-yl-spiro[1~{H}-indole-3,3'-pyrrolidine]-1'-carboxylate | | Authors: | Zhang, Z, Zhou, H. | | Deposit date: | 2019-06-16 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of novel liver X receptor inverse agonists as lipogenesis inhibitors.
Eur.J.Med.Chem., 206, 2020
|
|
8WX1
 
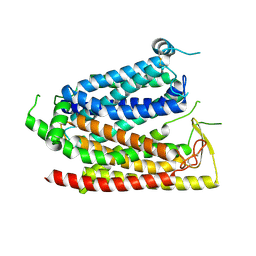 | |
8Y2M
 
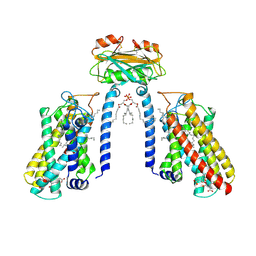 | | Cryo-EM structure of the FB1-bound Lac1-Lip1 complex | | Descriptor: | (2~{R})-2-[2-[(5~{R},6~{R},7~{S},9~{S},11~{R},16~{R},18~{S},19~{S})-19-azanyl-6-[(3~{R})-3-carboxy-5-oxidanyl-5-oxidanylidene-pentanoyl]oxy-5,9-dimethyl-11,16,18-tris(oxidanyl)icosan-7-yl]oxy-2-oxidanylidene-ethyl]butanedioic acid, (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, Ceramide synthase LAC1, ... | | Authors: | Xie, T, Zhang, Z, Fang, Q, Gong, X. | | Deposit date: | 2024-01-26 | | Release date: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Mechanism of ceramide synthase inhibition by fumonisin B 1.
Structure, 32, 2024
|
|
8Y2N
 
 | | Cryo-EM structure of the apo Lac1-Lip1 complex | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, Ceramide synthase LAC1, Ceramide synthase subunit LIP1 | | Authors: | Xie, T, Zhang, Z, Fang, Q, Gong, X. | | Deposit date: | 2024-01-26 | | Release date: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Mechanism of ceramide synthase inhibition by fumonisin B 1.
Structure, 32, 2024
|
|
6K0Z
 
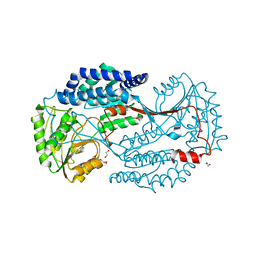 | | Substrate bound state of Staphylococcus Aureus AldH | | Descriptor: | 1,2-ETHANEDIOL, Aldehyde dehydrogenase, GLYCEROL | | Authors: | Zhang, Z, Tao, X. | | Deposit date: | 2019-05-08 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4967258 Å) | | Cite: | Structural Insight into the Substrate Gating Mechanism by Staphylococcus aureus Aldehyde Dehydrogenase
CCS Chemistry, 2, 2020
|
|
7E7X
 
 | |
2NCA
 
 | |
6LVY
 
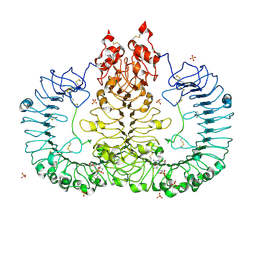 | | Crystal structure of TLR7/Cpd-2 (SM-360320) complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-azanyl-2-(2-methoxyethoxy)-9-(phenylmethyl)-7H-purin-8-one, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2020-02-06 | | Release date: | 2020-11-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis reveals TLR7 dynamics underlying antagonism.
Nat Commun, 11, 2020
|
|
6LW0
 
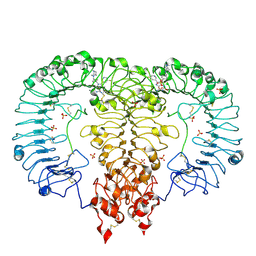 | | Crystal structure of TLR7/Cpd-6 (DSR-139293) complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-ethoxy-8-(5-fluoranylpyridin-3-yl)-9-[[4-[[(1S,4S)-5-methyl-2,5-diazabicyclo[2.2.1]heptan-2-yl]methyl]phenyl]methyl]purin-6-amine, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2020-02-06 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis reveals TLR7 dynamics underlying antagonism.
Nat Commun, 11, 2020
|
|
6LW1
 
 | | Cryo-EM structure of TLR7/Cpd-7 (DSR-139970) complex in open form | | Descriptor: | 2-ethoxy-8-(5-fluoranylpyridin-3-yl)-6-methyl-9-[[4-[[(1S,4S)-5-methyl-2,5-diazabicyclo[2.2.1]heptan-2-yl]methyl]phenyl]methyl]purine, Toll-like receptor 7 | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2020-02-06 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural analysis reveals TLR7 dynamics underlying antagonism.
Nat Commun, 11, 2020
|
|
7Y4F
 
 | | bacterial DPP4 | | Descriptor: | Dipeptidyl peptidase IV | | Authors: | Hang, J, Jiang, C, Wang, K, Zhang, Z, Guo, F, Liu, J, Wang, G, Lei, X, Gonzalez, F, Qiao, J. | | Deposit date: | 2022-06-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.918 Å) | | Cite: | Microbial-host-isozyme analyses reveal microbial DPP4 as a potential antidiabetic target.
Science, 381, 2023
|
|
7Y4G
 
 | | sit-bound btDPP4 | | Descriptor: | (2R)-4-OXO-4-[3-(TRIFLUOROMETHYL)-5,6-DIHYDRO[1,2,4]TRIAZOLO[4,3-A]PYRAZIN-7(8H)-YL]-1-(2,4,5-TRIFLUOROPHENYL)BUTAN-2-A MINE, btDPP4 | | Authors: | Hang, J, Jiang, C, Wang, K, Zhang, Z, Guo, F, Liu, J, Wang, G, Lei, X, Gonzalez, F, Qiao, J. | | Deposit date: | 2022-06-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Microbial-host-isozyme analyses reveal microbial DPP4 as a potential antidiabetic target.
Science, 381, 2023
|
|
8HAY
 
 | | d4-bound btDPP4 | | Descriptor: | (1~{R})-1-[[4-[5-[[(1~{R})-6,7-dimethoxy-2-methyl-3,4-dihydro-1~{H}-isoquinolin-1-yl]methyl]-2-methoxy-phenoxy]phenyl]methyl]-6,7-dimethoxy-2-methyl-3,4-dihydro-1~{H}-isoquinoline, btDPP4 | | Authors: | Hang, J, Jiang, C, Wang, K, Zhang, Z, Guo, F, Liu, J, Wang, G, Lei, X, Gonzalez, F, Qiao, J. | | Deposit date: | 2022-10-27 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Microbial-host-isozyme analyses reveal microbial DPP4 as a potential antidiabetic target.
Science, 381, 2023
|
|
6QLE
 
 | | Structure of inner kinetochore CCAN complex | | Descriptor: | Central kinetochore subunit CTF3,Inner kinetochore subunit CTF3,Central kinetochore subunit CTF3,Inner kinetochore subunit CTF3, Central kinetochore subunit MCM16,Central kinetochore subunit MCM16,Inner kinetochore subunit MCM16,Mcm16p, Inner kinetochore subunit AME1,Inner kinetochore subunit AME1,Inner kinetochore subunit AME1,Inner kinetochore subunit AME1, ... | | Authors: | Yan, K, Yang, J, Zhang, Z, McLaughlin, S.H, Chang, L, Fasci, D, Heck, A.J.R, Barford, D. | | Deposit date: | 2019-01-31 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structure of the inner kinetochore CCAN complex assembled onto a centromeric nucleosome.
Nature, 574, 2019
|
|
6QLF
 
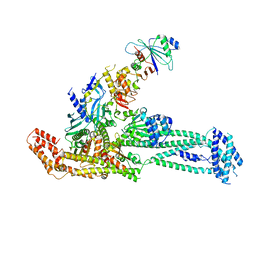 | | Structure of inner kinetochore CCAN complex with mask1 | | Descriptor: | Inner kinetochore subunit AME1, Inner kinetochore subunit CHL4, Inner kinetochore subunit CTF19, ... | | Authors: | Yan, K, Yang, J, Zhang, Z, McLaughlin, S.H, Chang, L, Fasci, D, Heck, A.J.R, Barford, D. | | Deposit date: | 2019-01-31 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure of the inner kinetochore CCAN complex assembled onto a centromeric nucleosome.
Nature, 574, 2019
|
|
