3WMB
 
 | | Crystal structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with naphthalimide derivative Q1 | | Descriptor: | 2-(2-{[(5-methyl-1,3,4-thiadiazol-2-yl)methyl]amino}ethyl)-1H-benzo[de]isoquinoline-1,3(2H)-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-hexosaminidase | | Authors: | Liu, T, Zhou, Y, Chen, L, Yang, Q. | | Deposit date: | 2013-11-16 | | Release date: | 2014-11-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A crystal structure-guided rational design switching non-carbohydrate inhibitors' specificity between two beta-GlcNAcase homologs
Sci Rep, 4, 2014
|
|
3X21
 
 | | Crystal structure of Escherichia coli nitroreductase NfsB mutant T41L/N71S/F124W | | Descriptor: | FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Bai, J, Yang, J, Zhou, Y, Yang, Q. | | Deposit date: | 2014-12-06 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Altering the regioselectivity of a nitroreductase in the synthesis of arylhydroxylamines by structure-based engineering.
Chembiochem, 16, 2015
|
|
3WZF
 
 | | Crystal structure of human cytoplasmic aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, cytoplasmic | | Authors: | Jiang, X, Chang, H, Zhou, Y, Chen, L, Yang, Q. | | Deposit date: | 2014-09-24 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.991 Å) | | Cite: | Recombinant expression, purification and Preliminary crystallographic studies of human cytoplasmic aspartate aminotransferase
To be Published
|
|
3X22
 
 | | Crystal structure of Escherichia coli nitroreductase NfsB mutant N71S/F123A/F124W | | Descriptor: | FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Bai, J, Yang, J, Zhou, Y, Yang, Q. | | Deposit date: | 2014-12-06 | | Release date: | 2015-11-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural basis of Escherichia coli nitroreductase NfsB triple mutants engineered for improved activity and regioselectivity toward the prodrug CB1954
PROCESS BIOCHEM, 50, 2015
|
|
7W0V
 
 | | C4'-SCF3-DT modifeid DNA-DNA duplex | | Descriptor: | DNA (5'-D(*CP*CP*AP*TP*(DSW)P*AP*TP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*TP*GP*G)-3') | | Authors: | Li, Q, Trajkovski, M, Fan, C, Chen, J, Zhou, Y, Lu, K, Li, H, Su, X, Xi, Z, Plavec, J, Zhou, C. | | Deposit date: | 2021-11-18 | | Release date: | 2022-11-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 4'-SCF 3 -Labeling Constitutes a Sensitive 19 F NMR Probe for Characterization of Interactions in the Minor Groove of DNA.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
2GCQ
 
 | |
7XSV
 
 | | Crystal Structures of PIM1 in Complex with Macrocyclic Compound H3 | | Descriptor: | 8-Methyl-2,5,20-trioxa-8,13,17-triazatetracyclo[11.10.2.014,19.021,25]pentacosa-1(24),14(19),15,17,21(25),22-hexaene, Serine/threonine-protein kinase pim-1 | | Authors: | Shen, C, Xie, Y, Ren, X, Zhou, Y, Niu, H. | | Deposit date: | 2022-05-15 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Design, synthesis, and bioactivity evaluation of macrocyclic benzo[b]pyrido[4,3-e][1,4]oxazine derivatives as novel Pim-1 kinase inhibitors.
Bioorg.Med.Chem.Lett., 72, 2022
|
|
1JVM
 
 | |
3S96
 
 | | Crystal structure of 3B5H10 | | Descriptor: | 3B5H10 FAB heavy chain, 3B5H10 FAB light chain | | Authors: | Weisgraber, K, Peters-Libeu, C, Rutenber, E, Newhouse, Y, Finkbeiner, S. | | Deposit date: | 2011-05-31 | | Release date: | 2012-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Disease-associated polyglutamine stretches in monomeric huntingtin adopt a compact structure.
J.Mol.Biol., 421, 2012
|
|
7VRG
 
 | | Crystal structure of chitinase-h from O. furnacalis in complex with Lynamicin B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase, ... | | Authors: | Lu, Q, Liu, T, Zhou, Y, Yang, Q. | | Deposit date: | 2021-10-22 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Lynamicin B is a Potential Pesticide by Acting as a Lepidoptera-Exclusive Chitinase Inhibitor.
J.Agric.Food Chem., 69, 2021
|
|
1C1J
 
 | | STRUCTURE OF CADMIUM-SUBSTITUTED PHOSPHOLIPASE A2 FROM AGKISTRONDON HALYS PALLAS AT 2.8 ANGSTROMS RESOLUTION | | Descriptor: | BASIC PHOSPHOLIPASE A2, CADMIUM ION, octyl beta-D-glucopyranoside | | Authors: | Zhang, H.-l, Zhang, Y.-q, Song, S.-y, Zhou, Y, Lin, Z.-j. | | Deposit date: | 1999-07-22 | | Release date: | 2002-07-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Cadmium-substituted Phospholipase A2 from Agkistrodon halys
Pallas at 2.8 Angstroms Resolution
Protein Pept.Lett., 6, 1999
|
|
7YFN
 
 | | Core module of the NuA4 complex in S. cerevisiae | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ARP4 isoform 1, Actin, ... | | Authors: | Ji, L.T, Zhao, L.X, Xu, K, Gao, H.H, Zhou, Y, Kornberg, R.D, Zhang, H.Q. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the NuA4 histone acetyltransferase complex.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7YFP
 
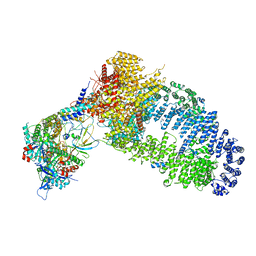 | | The NuA4 histone acetyltransferase complex from S. cerevisiae | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ARP4 isoform 1, Actin, ... | | Authors: | Ji, L.T, Zhao, L.X, Xu, K, Gao, H.H, Zhou, Y, Kornberg, R.D, Zhang, H.Q. | | Deposit date: | 2022-07-08 | | Release date: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of the NuA4 histone acetyltransferase complex.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1BK9
 
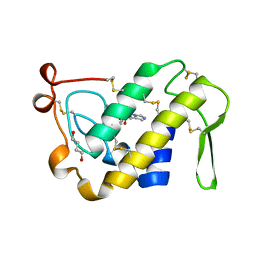 | | PHOSPHOLIPASE A2 MODIFIED BY PBPB | | Descriptor: | 1,4-BUTANEDIOL, CALCIUM ION, PHOSPHOLIPASE A2, ... | | Authors: | Zhao, H, Tang, L, Wang, X, Lin, Z, Zhou, Y. | | Deposit date: | 1998-07-16 | | Release date: | 1999-03-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a snake venom phospholipase A2 modified by p-bromo-phenacyl-bromide.
Toxicon, 36, 1998
|
|
1CXV
 
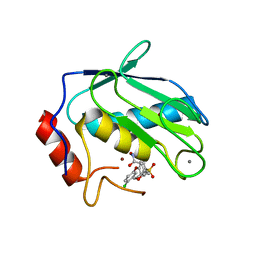 | | STRUCTURE OF RECOMBINANT MOUSE COLLAGENASE-3 (MMP-13) | | Descriptor: | 2-{4-[4-(4-CHLORO-PHENOXY)-BENZENESULFONYL]-TETRAHYDRO-PYRAN-4-YL}-N-HYDROXY-ACETAMIDE, CALCIUM ION, PROTEIN (COLLAGENASE-3), ... | | Authors: | Botos, I, Meyer, E, Swanson, S.M, Lemaitre, V, Eeckhout, Y, Meyer, E.F. | | Deposit date: | 1999-08-30 | | Release date: | 2000-08-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of recombinant mouse collagenase-3 (MMP-13).
J.Mol.Biol., 292, 1999
|
|
8W6R
 
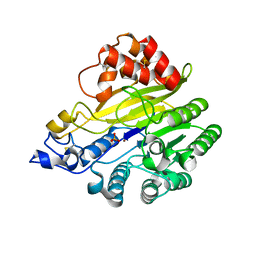 | | murine SMPDL3A bound to sulfate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acid sphingomyelinase-like phosphodiesterase 3a, ... | | Authors: | Zhang, C, Liu, P, Fan, S. | | Deposit date: | 2023-08-29 | | Release date: | 2023-12-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | SMPDL3A is a cGAMP-degrading enzyme induced by LXR-mediated lipid metabolism to restrict cGAS-STING DNA sensing.
Immunity, 56, 2023
|
|
5ZUU
 
 | | Crystal structure of AtCPSF30 YTH domain in complex with 10mer m6A-modified RNA | | Descriptor: | 30-kDa cleavage and polyadenylation specificity factor 30, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Wu, B, Nie, H, Li, S, Patel, D.J. | | Deposit date: | 2018-05-08 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | CPSF30-L-mediated recognition of mRNA m6A modification controls alternative polyadenylation of nitrate signaling-related gene transcripts in Arabidopsis.
Mol Plant, 2021
|
|
7WR3
 
 | | Crystal structure of MBP-fused OspC3 in complex with calmodulin | | Descriptor: | Calmodulin-1, MBP-fused OspC3, NICOTINAMIDE, ... | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR5
 
 | | Crystal structure of OspC3-calmodulin-caspase-4 complex binding with 2'-aF-NAD+ | | Descriptor: | Calmodulin-1, Caspase-4, OspC3, ... | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR4
 
 | | Crystal structure of OspC3-calmodulin-caspase-4 complex | | Descriptor: | Calmodulin-1, Caspase-4, OspC3 | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR0
 
 | | P32 of caspase-4 C258A mutant | | Descriptor: | Caspase-4 | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR6
 
 | | Crystal structure of ADP-riboxanated caspase-4 in complex with Af1521 | | Descriptor: | ADP-ribose glycohydrolase AF_1521, Caspase-4, [[(3~{a}~{S},5~{R},6~{R},6~{a}~{R})-2-azanylidene-3-[(4~{R})-4-azanyl-5-oxidanylidene-pentyl]-6-oxidanyl-3~{a},5,6,6~{a}-tetrahydrofuro[2,3-d][1,3]oxazol-5-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR1
 
 | | P32 of caspase-4 C258A mutant in complex with OspC3 C-terminal ankyrin-repeat domain | | Descriptor: | Caspase-4, OspC3 | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR2
 
 | |
2OY0
 
 | | Crystal structure of the West Nile virus methyltransferase | | Descriptor: | Methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Li, H.M, Zhao, Y.W, Guo, Y, Shi, P.Y. | | Deposit date: | 2007-02-21 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and Function of Flavivirus NS5 Methyltransferase.
J.Virol., 81, 2007
|
|
