5J36
 
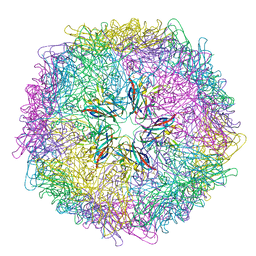 | | Crystal structure of 60-mer BFDV Capsid Protein | | Descriptor: | Beak and feather disease virus capsid protein, PHOSPHATE ION | | Authors: | Sarker, S, Raidal, S, Aragao, D, Forwood, J.K. | | Deposit date: | 2016-03-30 | | Release date: | 2016-05-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural insights into the assembly and regulation of distinct viral capsid complexes.
Nat Commun, 7, 2016
|
|
6RNJ
 
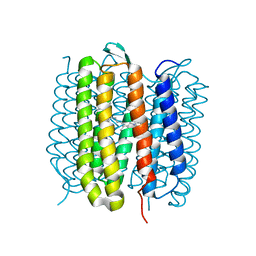 | | TR-SMX closed state structure (0-5ms) of bacteriorhodopsin | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Weinert, T, Skopintsev, P, James, D, Kekilli, D, Furrer, F, Bruenle, S, Mous, S, Nogly, P, Standfuss, J. | | Deposit date: | 2019-05-08 | | Release date: | 2019-07-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Proton uptake mechanism in bacteriorhodopsin captured by serial synchrotron crystallography.
Science, 365, 2019
|
|
6HL1
 
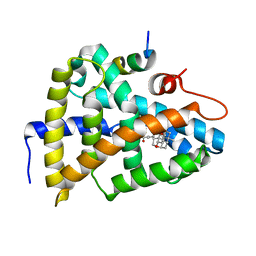 | | Crystal Structure of Farnesoid X receptor (FXR) with bound NCoA-2 peptide and CDCA | | Descriptor: | Bile acid receptor, CHENODEOXYCHOLIC ACID, NCoA-2 peptide (Nuclear receptor coactivator 2), ... | | Authors: | Kudlinzki, D, Merk, D, Linhard, V.L, Saxena, K, Schubert-Zsilavecz, M, Schwalbe, H. | | Deposit date: | 2018-09-10 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Molecular tuning of farnesoid X receptor partial agonism.
Nat Commun, 10, 2019
|
|
6HL0
 
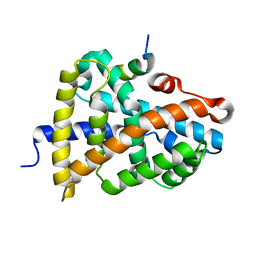 | | Crystal Structure of Farnesoid X receptor (FXR) with bound NCoA-2 peptide | | Descriptor: | Bile acid receptor, NCoA-2 peptide (Nuclear receptor coactivator 2), LYS-GLU-ASN-ALA-LEU-LEU-ARG-TYR-LEU-LEU-ASP-LYS-ASP | | Authors: | Kudlinzki, D, Merk, D, Linhard, V.L, Saxena, K, Schubert-Zsilavecz, M, Schwalbe, H. | | Deposit date: | 2018-09-10 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Molecular tuning of farnesoid X receptor partial agonism.
Nat Commun, 10, 2019
|
|
5J09
 
 | |
9JBP
 
 | | Cryo-EM structure of human SOD1 (C6A/C111A) amyloid filament | | Descriptor: | Superoxide dismutase [Cu-Zn], Unassigned poly-alanine model | | Authors: | Baek, Y, Kim, H, Lee, D, Kim, D, Jo, E, Roh, S.-H, Ha, N.-C. | | Deposit date: | 2024-08-27 | | Release date: | 2025-01-15 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural insights into the role of reduced cysteine residues in SOD1 amyloid filament formation.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9JBO
 
 | | Cryo-EM structure of human SOD1 (WT) amyloid filament | | Descriptor: | Superoxide dismutase [Cu-Zn] | | Authors: | Baek, Y, Kim, H, Lee, D, Kim, D, Jo, E, Roh, S.-H, Ha, N.-C. | | Deposit date: | 2024-08-27 | | Release date: | 2025-01-15 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural insights into the role of reduced cysteine residues in SOD1 amyloid filament formation.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
1EPH
 
 | |
5I9X
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with bosutinib (SKI-606) | | Descriptor: | 1,2-ETHANEDIOL, 4-[(2,4-dichloro-5-methoxyphenyl)amino]-6-methoxy-7-[3-(4-methylpiperazin-1-yl)propoxy]quinoline-3-carbonitrile, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Linhard, V.L, Gande, S.L, Sreeramulu, S, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2016-02-21 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.427 Å) | | Cite: | Chemical Proteomics and Structural Biology Define EPHA2 Inhibition by Clinical Kinase Drugs.
ACS Chem. Biol., 11, 2016
|
|
6SND
 
 | | crystal structure of LN01 Fab in complex with an HIV-1 gp41 peptide | | Descriptor: | 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Caillat, C, Pinto, D, Corti, D, Fenwick, C, Pantaleo, G, Weissenhorn, W. | | Deposit date: | 2019-08-23 | | Release date: | 2019-11-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis for Broad HIV-1 Neutralization by the MPER-Specific Human Broadly Neutralizing Antibody LN01.
Cell Host Microbe, 26, 2019
|
|
5MMP
 
 | | E. coli DNA Gyrase B 24 kDa ATPase domain in complex with 1-ethyl-3-[5-pyridin-4-yl-8-(pyridin-3-ylamino)-isoquinolin-3-yl]-urea | | Descriptor: | 1-ethyl-3-[5-pyridin-4-yl-8-(pyridin-3-ylamino)isoquinolin-3-yl]urea, DNA gyrase subunit B | | Authors: | Panchaud, P, Bruyere, T, Blumstein, A.-C, Bur, D, Chambovey, A, Ertel, E.A, Gude, M, Hubschwerlen, C, Jacob, L, Kimmerlin, T, Pfeifer, T, Prade, L, Seiler, P, Ritz, D, Rueedi, G. | | Deposit date: | 2016-12-12 | | Release date: | 2017-04-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery and Optimization of Isoquinoline Ethyl Ureas as Antibacterial Agents.
J. Med. Chem., 60, 2017
|
|
6HBJ
 
 | | Echovirus 18 empty particle | | Descriptor: | Viral protein 1, Viral protein 2, Viral protein 3 | | Authors: | Buchta, D, Fuzik, T, Hrebik, D, Levdansky, Y, Moravcova, J, Plevka, P. | | Deposit date: | 2018-08-10 | | Release date: | 2019-03-20 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Enterovirus particles expel capsid pentamers to enable genome release.
Nat Commun, 10, 2019
|
|
7B5F
 
 | | Structure of echovirus 18 in complex with neonatal Fc receptor | | Descriptor: | Beta-2-microglobulin, Echovirus 18 viral protein 1, Echovirus 18 viral protein 2, ... | | Authors: | Buchta, D, Levdansky, Y, Fuzik, T, Mukhamedova, L, Moravcova, J, Hrebik, D, Andersen, J.T, Plevka, P. | | Deposit date: | 2020-12-03 | | Release date: | 2022-01-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of echovirus 18 in complex with neonatal Fc receptor
To Be Published
|
|
9GS7
 
 | |
9GS5
 
 | |
5VZ3
 
 | |
6HVB
 
 | | NMR structure of Urotensin Peptide Asp-c[Cys-Phe-(N-Me)Trp-Lys-Tyr-Cys]-Val in SDS solution | | Descriptor: | Urotensin-2 | | Authors: | Brancaccio, D, Carotenuto, A, Merlino, F, Billard, E, Yousif, A.M, Di Maro, S, Abate, L, Bellavita, R, D'Emmanuele di Villa Bianca, R, Santicioli, P, Marinelli, L, Novellino, E, Hebert, T.E, Lubell, W.D, Chatenet, D, Grieco, P. | | Deposit date: | 2018-10-10 | | Release date: | 2019-01-16 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Functional Selectivity Revealed by N-Methylation Scanning of Human Urotensin II and Related Peptides.
J.Med.Chem., 62, 2019
|
|
4NC3
 
 | | Crystal structure of the 5-HT2B receptor solved using serial femtosecond crystallography in lipidic cubic phase. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHOLESTEROL, ... | | Authors: | Liu, W, Wacker, D, Gati, C, Han, G.W, James, D, Wang, D, Nelson, G, Weierstall, U, Katritch, V, Barty, A, Zatsepin, N.A, Li, D, Messerschmidt, M, Boutet, S, Williams, G.J, Koglin, J.E, Seibert, M.M, Wang, C, Shah, S.T.A, Basu, S, Fromme, R, Kupitz, C, Rendek, K.N, Grotjohann, I, Fromme, P, Kirian, R.A, Beyerlein, K.R, White, T.A, Chapman, H.N, Caffrey, M, Spence, J.C.H, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Serial femtosecond crystallography of G protein-coupled receptors.
Science, 342, 2013
|
|
1J6Y
 
 | | Solution structure of Pin1At from Arabidopsis thaliana | | Descriptor: | peptidyl-prolyl cis-trans isomerase | | Authors: | Landrieu, I, Wieruszeski, J.M, Wintjens, R, Inze, D, Lippens, G. | | Deposit date: | 2001-05-15 | | Release date: | 2002-08-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the
Single-domain Prolyl Cis/Trans
Isomerase PIN1At from Arabidopsis thaliana
J.Mol.Biol., 320, 2002
|
|
6I7D
 
 | | Plasmodium falciparum Myosin A, post-rigor and rigor-like states | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Myosin-A | | Authors: | Robert-Paganin, J, Auguin, D, Moussaoui, D, Jousset, G, Baum, J, Trybus, K.M, Houdusse, A. | | Deposit date: | 2018-11-16 | | Release date: | 2019-08-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Plasmodium myosin A drives parasite invasion by an atypical force generating mechanism.
Nat Commun, 10, 2019
|
|
5VOS
 
 | | VGSNKGAIIGL from Amyloid Beta determined by MicroED | | Descriptor: | Amyloid beta A4 protein | | Authors: | Rodriguez, J.A, Sawaya, M.R, Cascio, D, Eisenberg, D.S, Griner, S.L, Gonen, T. | | Deposit date: | 2017-05-03 | | Release date: | 2018-01-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.42 Å) | | Cite: | Common fibrillar spines of amyloid-beta and human islet amyloid polypeptide revealed by microelectron diffraction and structure-based inhibitors.
J. Biol. Chem., 293, 2018
|
|
5LOQ
 
 | | Structure of coproheme bound HemQ from Listeria monocytogenes | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, Putative heme-dependent peroxidase lmo2113, ... | | Authors: | Puehringer, D, Mlynek, G, Hofbauer, S, Djinovic-Carugo, K, Obinger, C. | | Deposit date: | 2016-08-09 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Hydrogen peroxide-mediated conversion of coproheme to heme b by HemQ-lessons from the first crystal structure and kinetic studies.
FEBS J., 283, 2016
|
|
3FE5
 
 | | Crystal structure of 3-hydroxyanthranilate 3,4-dioxygenase from bovine kidney | | Descriptor: | 3-hydroxyanthranilate 3,4-dioxygenase, FE (III) ION | | Authors: | Dilovic, I, Gliubich, F, Malpeli, G, Zanotti, G, Matkovic-Calogovic, D. | | Deposit date: | 2008-11-27 | | Release date: | 2009-06-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of bovine 3-hydroxyanthranilate 3,4-dioxygenase.
Biopolymers, 2009
|
|
5WRN
 
 | |
1CGL
 
 | | Structure of the catalytic domain of fibroblast collagenase complexed with an inhibitor | | Descriptor: | CALCIUM ION, FIBROBLAST COLLAGENASE, N-[(1S)-3-{[(benzyloxy)carbonyl]amino}-1-carboxypropyl]-L-leucyl-N-(2-morpholin-4-ylethyl)-L-phenylalaninamide, ... | | Authors: | Lovejoy, B, Cleasby, A, Hassell, A.M, Longley, K, Luther, M.A, Weigl, D, Mcgeehan, G, Mcelroy, A.B, Drewry, D, Lambert, M.H, Jordan, S.R. | | Deposit date: | 1993-11-17 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the catalytic domain of fibroblast collagenase complexed with an inhibitor.
Science, 263, 1994
|
|
