1BID
 
 | |
1AOB
 
 | | E. COLI THYMIDYLATE SYNTHASE COMPLEXED WITH DDURD | | Descriptor: | 2'-5'DIDEOXYURIDINE, FORMIC ACID, PHOSPHATE ION, ... | | Authors: | Stout, T.J, Sage, C.R, Stroud, R.M. | | Deposit date: | 1997-06-30 | | Release date: | 1998-07-01 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The additivity of substrate fragments in enzyme-ligand binding.
Structure, 6, 1998
|
|
1BDU
 
 | |
1BKO
 
 | |
1BSF
 
 | |
1BKP
 
 | |
1BSP
 
 | | THERMOSTABLE THYMIDYLATE SYNTHASE A FROM BACILLUS SUBTILIS | | Descriptor: | PHOSPHATE ION, THYMIDYLATE SYNTHASE A | | Authors: | Stout, T.J, Schellenberger, U, Santi, D.V, Stroud, R.M. | | Deposit date: | 1998-07-09 | | Release date: | 1999-02-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of a unique thermal-stable thymidylate synthase from Bacillus subtilis.
Biochemistry, 37, 1998
|
|
1AN5
 
 | | E. COLI THYMIDYLATE SYNTHASE IN COMPLEX WITH CB3717 | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, PHOSPHATE ION, THYMIDYLATE SYNTHASE | | Authors: | Stout, T.J, Sage, C.R, Stroud, R.M. | | Deposit date: | 1997-06-26 | | Release date: | 1998-07-01 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The additivity of substrate fragments in enzyme-ligand binding.
Structure, 6, 1998
|
|
1AID
 
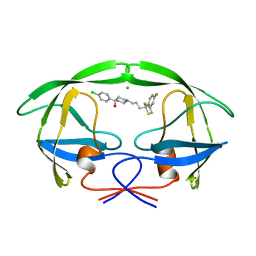 | | STRUCTURE OF A NON-PEPTIDE INHIBITOR COMPLEXED WITH HIV-1 PROTEASE: DEVELOPING A CYCLE OF STRUCTURE-BASED DRUG DESIGN | | Descriptor: | 4-(4-CHLORO-PHENYL)-1-{3-[2-(4-FLUORO-PHENYL)-[1,3]DITHIOLAN-2-YL]-PROPYL}-PIPERIDIN-4-OL, CHLORIDE ION, HUMAN IMMUNODEFICIENCY VIRUS PROTEASE | | Authors: | Rutenber, E.E, Fauman, E.B, Keenan, R.J, Stroud, R.M. | | Deposit date: | 1997-04-16 | | Release date: | 1997-10-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a non-peptide inhibitor complexed with HIV-1 protease. Developing a cycle of structure-based drug design.
J.Biol.Chem., 268, 1993
|
|
1CI7
 
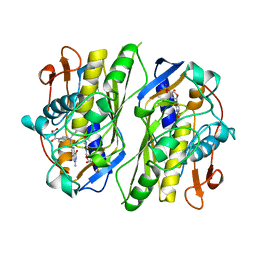 | | TERNARY COMPLEX OF THYMIDYLATE SYNTHASE FROM PNEUMOCYSTIS CARINII | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, PROTEIN (THYMIDYLATE SYNTHASE) | | Authors: | Anderson, A.C, O'Neil, R.H, Delano, W.L, Stroud, R.M. | | Deposit date: | 1999-04-08 | | Release date: | 2000-04-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structural mechanism for half-the-sites reactivity in an enzyme, thymidylate synthase, involves a relay of changes between subunits.
Biochemistry, 38, 1999
|
|
7JX1
 
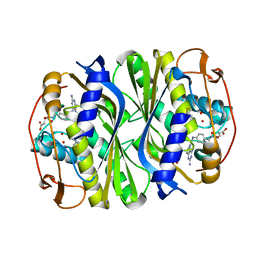 | | E. coli TSase complex with a bi-substrate reaction intermediate analog | | Descriptor: | (2S)-2-({4-[({(6R)-2,4-diamino-5-[(1-{(2R,4S,5R)-4-hydroxy-5-[(phosphonooxy)methyl]tetrahydrofuran-2-yl}-2,4-dioxo-1,2,3,4-tetrahydropyrimidin-5-yl)methyl]-5,6,7,8-tetrahydropyrido[3,2-d]pyrimidin-6-yl}methyl)amino]benzoyl}amino)pentanedioic acid (non-preferred name), N-(4-{[(2,4-diamino-7,8-dihydropyrido[3,2-d]pyrimidin-6-yl)methyl]amino}benzene-1-carbonyl)-L-glutamic acid, PHOSPHATE ION, ... | | Authors: | Finer-Moore, J, Kholodar, S.A, Stroud, R.M, Kohen, A. | | Deposit date: | 2020-08-26 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Caught in Action: X-ray Structure of Thymidylate Synthase with Noncovalent Intermediate Analog.
Biochemistry, 60, 2021
|
|
7JXF
 
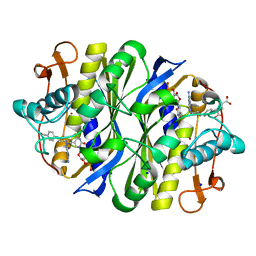 | | E. coli TSase complex with a bi-substrate reaction intermediate diastereomer analog | | Descriptor: | (2S)-2-({4-[({(6S)-2,4-diamino-5-[(1-{(2R,4S,5R)-4-hydroxy-5-[(phosphonooxy)methyl]tetrahydrofuran-2-yl}-2,4-dioxo-1,2,3,4-tetrahydropyrimidin-5-yl)methyl]-5,6,7,8-tetrahydropyrido[3,2-d]pyrimidin-6-yl}methyl)amino]benzoyl}amino)pentanedioic acid (non-preferred name), 5-HYDROXYMETHYLURIDINE-2'-DEOXY-5'-MONOPHOSPHATE, N-[4-({[(6S)-2,4-diamino-5,6,7,8-tetrahydropyrido[3,2-d]pyrimidin-6-yl]methyl}amino)benzene-1-carbonyl]-L-glutamic acid, ... | | Authors: | Finer-Moore, J, Kholodar, S.A, Stroud, R.M, Kohen, A, Moliner, V, Swiderek, K. | | Deposit date: | 2020-08-27 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Caught in Action: X-ray Structure of Thymidylate Synthase with Noncovalent Intermediate Analog.
Biochemistry, 60, 2021
|
|
7KTX
 
 | | Cryo-EM structure of Saccharomyces cerevisiae ER membrane protein complex bound to a Fab in DDM detergent | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ER membrane protein complex subunit 1, ER membrane protein complex subunit 2, ... | | Authors: | Miller-Vedam, L.E, Schirle Oakdale, N.S, Braeuning, B, Boydston, E.A, Sevillano, N, Popova, K.D, Bonnar, J.L, Shurtleff, M.J, Prabu, J.R, Stroud, R.M, Craik, C.S, Schulman, B.A, Weissman, J.S, Frost, A. | | Deposit date: | 2020-11-24 | | Release date: | 2020-12-02 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural and mechanistic basis of the EMC-dependent biogenesis of distinct transmembrane clients.
Elife, 9, 2020
|
|
7KP4
 
 | |
7KRA
 
 | | Cryo-EM structure of Saccharomyces cerevisiae ER membrane protein complex bound to Fab-DH4 in lipid nanodiscs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ER membrane protein complex subunit 1, ER membrane protein complex subunit 2, ... | | Authors: | Miller-Vedam, L.E, Schirle Oakdale, N.S, Braeuning, B, Boydston, E.A, Sevillano, N, Popova, K.D, Bonnar, J.L, Shurtleff, M.J, Prabu, J.R, Stroud, R.M, Craik, C.S, Schulman, B.A, Weissman, J.S, Frost, A. | | Deposit date: | 2020-11-19 | | Release date: | 2020-12-02 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and mechanistic basis of the EMC-dependent biogenesis of distinct transmembrane clients.
Elife, 9, 2020
|
|
3UWL
 
 | | Crystal structure of Enteroccocus faecalis thymidylate synthase (EfTS) in complex with 5-formyl tetrahydrofolate | | Descriptor: | 1,2-ETHANEDIOL, N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, SULFATE ION, ... | | Authors: | Pozzi, C, Catalano, A, Cortesi, D, Luciani, R, Ferrari, S, Fritz, T, Costi, M.P, Mangani, S. | | Deposit date: | 2011-12-02 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The structure of Enterococcus faecalis thymidylate synthase provides clues about folate bacterial metabolism.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
7ADO
 
 | | Cryo-EM structure of human ER membrane protein complex in lipid nanodiscs | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ER membrane protein complex subunit 1, ... | | Authors: | Braeuning, B, Prabu, J.R, Miller-Vedam, L.E, Weissman, J.S, Frost, A, Schulman, B.A. | | Deposit date: | 2020-09-15 | | Release date: | 2020-12-02 | | Last modified: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural and mechanistic basis of the EMC-dependent biogenesis of distinct transmembrane clients.
Elife, 9, 2020
|
|
7ADP
 
 | | Cryo-EM structure of human ER membrane protein complex in GDN detergent | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ER membrane protein complex subunit 1, ER membrane protein complex subunit 10, ... | | Authors: | Braeuning, B, Prabu, J.R, Miller-Vedam, L.E, Weissman, J.S, Frost, A, Schulman, B.A. | | Deposit date: | 2020-09-15 | | Release date: | 2020-12-02 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural and mechanistic basis of the EMC-dependent biogenesis of distinct transmembrane clients.
Elife, 9, 2020
|
|
4TMS
 
 | |
1NTP
 
 | |
4ZE3
 
 | | Saccharomyces cerevisiae CYP51 (Lanosterol 14-alpha demethylase) Y140H mutant complexed with fluconazole | | Descriptor: | 2-(2,4-DIFLUOROPHENYL)-1,3-DI(1H-1,2,4-TRIAZOL-1-YL)PROPAN-2-OL, Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sagatova, A, Keniya, M.V, Wilson, R, Monk, B.C, Tyndall, J.D.A. | | Deposit date: | 2015-04-20 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Triazole resistance mediated by mutations of a conserved active site tyrosine in fungal lanosterol 14 alpha-demethylase.
Sci Rep, 6, 2016
|
|
3SDM
 
 | | Structure of oligomeric kinase/RNase Ire1 in complex with an oligonucleotide | | Descriptor: | Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Korennykh, A, Korostelev, A, Egea, P, Finer-Moore, J, Zhang, C, Stroud, R, Shokat, K, Walter, P. | | Deposit date: | 2011-06-09 | | Release date: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (6.6 Å) | | Cite: | Cofactor-mediated conformational control in the bifunctional kinase/RNase Ire1.
Bmc Biol., 9, 2011
|
|
7T4X
 
 | |
2ABX
 
 | |
3B7D
 
 | |
