8HD6
 
 | | The relaxed pre-Tet-S1 state of G264A mutated Tetrahymena group I intron with 6nt 3'/5'-exon and 2-aminopurine nucleoside | | Descriptor: | MAGNESIUM ION, SPERMIDINE, The relaxed pre-Tet-S1 state molecule of co-transcriptional folded G264A mutant Tetrahymena group I intron with 6nt 3'/5'-exon and 2-aminopurine nucleoside | | Authors: | Luo, B, Zhang, C, Ling, X, Mukherjee, S, Jia, G, Xie, J, Jia, X, Liu, L, Baulin, E.F, Luo, Y, Jiang, L, Dong, H, Wei, X, Bujnicki, J.M, Su, Z. | | Deposit date: | 2022-11-03 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Cryo-EM reveals dynamics of Tetrahymena group I intron self-splicing
Nat Catal, 2023
|
|
8HD7
 
 | | The intermediate pre-Tet-S1 state of G264A mutated Tetrahymena group I intron with 6nt 3'/5'-exon and 2-aminopurine nucleoside | | Descriptor: | MAGNESIUM ION, SPERMIDINE, The intermediate pre-Tet-S1 state molecule of co-transcriptional folded G264A mutant Tetrahymena group I intron with 6nt 3'/5'-exon and 2-aminopurine nucleoside | | Authors: | Luo, B, Zhang, C, Ling, X, Mukherjee, S, Jia, G, Xie, J, Jia, X, Liu, L, Baulin, E.F, Luo, Y, Jiang, L, Dong, H, Wei, X, Bujnicki, J.M, Su, Z. | | Deposit date: | 2022-11-03 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Cryo-EM reveals dynamics of Tetrahymena group I intron self-splicing
Nat Catal, 2023
|
|
5Z9Z
 
 | | The C-terminal RRM domain of Arabidopsis SMALL RNA DEGRADING NUCLEASE 1 (E329A/E330A/E332A) | | Descriptor: | CITRATE ANION, Small RNA degrading nuclease 1 | | Authors: | Chen, J, Liu, L, You, C, Gu, J, Ruan, W, Zhang, L, Cao, C, Gan, J, Huang, Y, Chen, X, Ma, J. | | Deposit date: | 2018-02-05 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Structural and biochemical insights into small RNA 3' end trimming by Arabidopsis SDN1.
Nat Commun, 9, 2018
|
|
4RIY
 
 | | Crystal structure of an EGFR/HER3 kinase domain heterodimer containing the cancer-associated HER3-E909G mutation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Epidermal growth factor receptor, MAGNESIUM ION, ... | | Authors: | Littlefield, P, Liu, L, Jura, N. | | Deposit date: | 2014-10-07 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.981 Å) | | Cite: | Structural analysis of the EGFR/HER3 heterodimer reveals the molecular basis for activating HER3 mutations.
Sci.Signal., 7, 2014
|
|
4RIX
 
 | | Crystal structure of an EGFR/HER3 kinase domain heterodimer containing the cancer-associated HER3-Q790R mutation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Epidermal growth factor receptor, MAGNESIUM ION, ... | | Authors: | Littlefield, P, Liu, L, Jura, N. | | Deposit date: | 2014-10-07 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural analysis of the EGFR/HER3 heterodimer reveals the molecular basis for activating HER3 mutations.
Sci.Signal., 7, 2014
|
|
5FFC
 
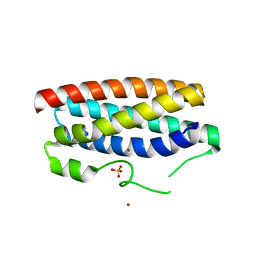 | | CopM in the Cu(II)-bound form | | Descriptor: | COPPER (II) ION, CopM, SULFATE ION | | Authors: | Zhao, S, Wang, X, Liu, L. | | Deposit date: | 2015-12-18 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | Structural basis for copper/silver binding by the Synechocystis metallochaperone CopM.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5FEJ
 
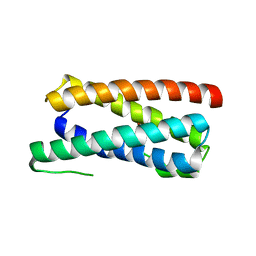 | | CopM in the Cu(I)-bound form | | Descriptor: | COPPER (I) ION, CopM | | Authors: | Zhao, S, Wang, X, Liu, L. | | Deposit date: | 2015-12-17 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for copper/silver binding by the Synechocystis metallochaperone CopM.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5FFA
 
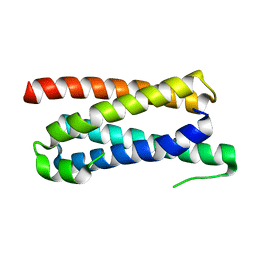 | |
5FFE
 
 | | CopM in the Ag-bound form (by soaking) | | Descriptor: | CopM, SILVER ION | | Authors: | Zhao, S, Wang, X, Liu, L. | | Deposit date: | 2015-12-18 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Structural basis for copper/silver binding by the Synechocystis metallochaperone CopM.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5FW5
 
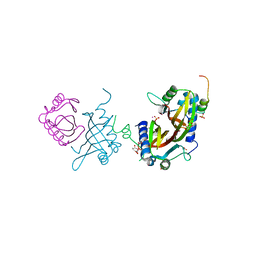 | | Crystal structure of human G3BP1 in complex with Semliki Forest Virus nsP3-25 comprising two FGDF motives | | Descriptor: | ACETATE ION, GLYCEROL, NON-STRUCTURAL PROTEIN 3, ... | | Authors: | Schulte, T, Liu, L, Panas, M.D, Thaa, B, Goette, B, Achour, A, McInerney, G.M. | | Deposit date: | 2016-02-12 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Combined structural, biochemical and cellular evidence demonstrates that both FGDF motifs in alphavirus nsP3 are required for efficient replication.
Open Biol, 6, 2016
|
|
5FFB
 
 | | CopM in the apo form | | Descriptor: | CopM | | Authors: | Zhao, S, Wang, X, Liu, L. | | Deposit date: | 2015-12-18 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Structural basis for copper/silver binding by the Synechocystis metallochaperone CopM.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
8GQP
 
 | | Complex of D-protein binder D-19437 and L-target L-Pep-1 | | Descriptor: | D-binder, L-pep1 | | Authors: | Liang, M.F, Li, S.C, Wang, T.Y, Liu, L, Lu, P.L. | | Deposit date: | 2022-08-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Accurate de novo design of heterochiral protein-protein interactions
Cell Res., 2024
|
|
5FFD
 
 | |
5H3E
 
 | | Crystal structure of mouse isocitrate dehydrogenases 2 K256Q mutant complexed with isocitrate | | Descriptor: | ISOCITRIC ACID, Isocitrate dehydrogenase [NADP], mitochondrial, ... | | Authors: | Xu, Y, Liu, L, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2016-10-23 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Studies on the regulatory mechanism of isocitrate dehydrogenase 2 using acetylation mimics
Sci Rep, 7, 2017
|
|
8WG5
 
 | | Cryo-EM structure of USP16 bound to H2AK119Ub nucleosome | | Descriptor: | DNA (147-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | Ai, H.S, He, Z.Z, Deng, Z.H, Liu, L. | | Deposit date: | 2023-09-20 | | Release date: | 2023-12-27 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural and mechanistic basis for nucleosomal H2AK119 deubiquitination by single-subunit deubiquitinase USP16.
Nat.Struct.Mol.Biol., 2024
|
|
5H3F
 
 | | Crystal structure of mouse isocitrate dehydrogenases 2 complexed with isocitrate | | Descriptor: | ISOCITRIC ACID, Isocitrate dehydrogenase [NADP], mitochondrial, ... | | Authors: | Xu, Y, Liu, L, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2016-10-23 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Studies on the regulatory mechanism of isocitrate dehydrogenase 2 using acetylation mimics
Sci Rep, 7, 2017
|
|
5GND
 
 | | Structure of Deg protease HhoA from Synechocystis sp. PCC 6803 | | Descriptor: | Putative serine protease HhoA, SODIUM ION, UNK-UNK-UNK-UNK-TRP, ... | | Authors: | Dong, W, Wang, J, Liu, L. | | Deposit date: | 2016-07-20 | | Release date: | 2016-09-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the zinc-bound HhoA protease from Synechocystis sp. PCC 6803
Febs Lett., 590, 2016
|
|
5HGZ
 
 | | Crystal structure of human Naa60 in complex with acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, MALONIC ACID, N-alpha-acetyltransferase 60 | | Authors: | Chen, J.Y, Liu, L, Yun, C.H. | | Deposit date: | 2016-01-09 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.383 Å) | | Cite: | Structure and function of human Naa60 (NatF), a Golgi-localized bi-functional acetyltransferase
Sci Rep, 6, 2016
|
|
5HH1
 
 | |
8HR1
 
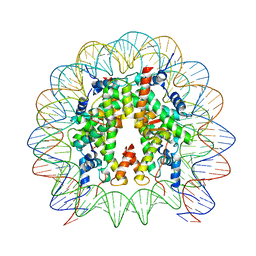 | | Cryo-EM structure of SSX1 bound to the unmodified nucleosome at a resolution of 3.02 angstrom | | Descriptor: | DNA (147-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | Zebin, T, Ai, H.S, Ziyu, X, Man, P, Liu, L. | | Deposit date: | 2022-12-14 | | Release date: | 2023-09-13 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Synovial sarcoma X breakpoint 1 protein uses a cryptic groove to selectively recognize H2AK119Ub nucleosomes.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HQY
 
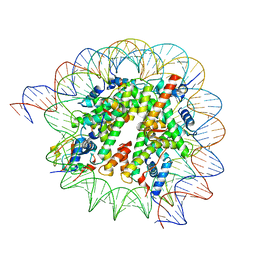 | | Cryo-EM structure of SSX1 bound to the H2AK119Ub nucleosome at a resolution of 3.05 angstrom | | Descriptor: | DNA (136-MER), DNA (137-MER), Histone H2A type 1-B/E, ... | | Authors: | Zebin, T, Ai, H.S, Ziyu, X, GuoChao, C, Man, P, Liu, L. | | Deposit date: | 2022-12-14 | | Release date: | 2023-09-27 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Synovial sarcoma X breakpoint 1 protein uses a cryptic groove to selectively recognize H2AK119Ub nucleosomes.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4GM8
 
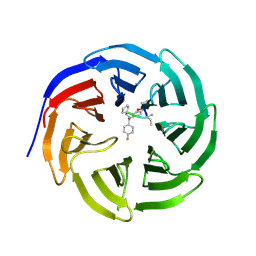 | | Crystal structure of human WD repeat domain 5 with compound MM-102 | | Descriptor: | MM-102, WD repeat-containing protein 5 | | Authors: | Karatas, H, Townsend, E.C, Chen, Y, Bernard, D, Cao, F, Liu, L, Lei, M, Dou, Y, Wang, S. | | Deposit date: | 2012-08-15 | | Release date: | 2013-07-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | High-affinity, small-molecule peptidomimetic inhibitors of MLL1/WDR5 protein-protein interaction.
J.Am.Chem.Soc., 135, 2013
|
|
4GHT
 
 | | Crystal structure of EV71 3C proteinase in complex with AG7088 | | Descriptor: | 3C proteinase, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | Authors: | Chen, C, Wu, C, Cai, Q, Li, N, Peng, X, Cai, Y, Yin, K, Chen, X, Wang, X, Zhang, R, Liu, L, Chen, S, Li, J, Lin, T. | | Deposit date: | 2012-08-08 | | Release date: | 2013-06-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structures of Enterovirus 71 3C proteinase (strain E2004104-TW-CDC) and its complex with rupintrivir
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4J4G
 
 | |
4J4E
 
 | |
