6KMX
 
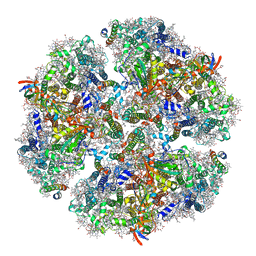 | | Structure of PSI from H. hongdechloris grown under far-red light condition | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Kato, K, Nagao, R, Shen, J.R, Miyazaki, N, Akita, F. | | Deposit date: | 2019-08-01 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Structural basis for the adaptation and function of chlorophyll f in photosystem I.
Nat Commun, 11, 2020
|
|
6KMW
 
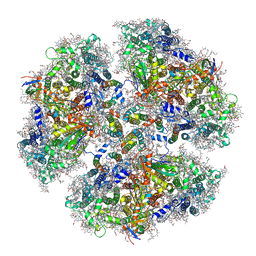 | | Structure of PSI from H. hongdechloris grown under white light condition | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Kato, K, Nagao, R, Shen, J.R, Miyazaki, N, Akita, F. | | Deposit date: | 2019-08-01 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Structural basis for the adaptation and function of chlorophyll f in photosystem I.
Nat Commun, 11, 2020
|
|
1YNR
 
 | | Crystal structure of the cytochrome c-552 from Hydrogenobacter thermophilus at 2.0 resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cytochrome c-552, HEME C, ... | | Authors: | Travaglini-Allocatelli, C, Gianni, S, Dubey, V.K, Borgia, A, Di Matteo, A, Bonivento, D, Cutruzzola, F, Bren, K.L, Brunori, M. | | Deposit date: | 2005-01-25 | | Release date: | 2005-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An Obligatory Intermediate in the Folding Pathway of Cytochrome c552 from Hydrogenobacter thermophilus
J.Biol.Chem., 280, 2005
|
|
8E8U
 
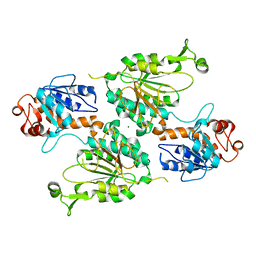 | | Structure of the LOR domain of human AASS | | Descriptor: | Alpha-aminoadipic semialdehyde synthase, mitochondrial, MAGNESIUM ION | | Authors: | Khamrui, S, Lazarus, M.B. | | Deposit date: | 2022-08-25 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Characterization and structure of the human lysine-2-oxoglutarate reductase domain, a novel therapeutic target for treatment of glutaric aciduria type 1.
Open Biology, 12, 2022
|
|
8E8V
 
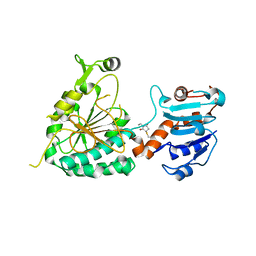 | |
8E8T
 
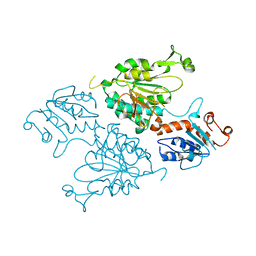 | | Structure of the short LOR domain of human AASS | | Descriptor: | Alpha-aminoadipic semialdehyde synthase, mitochondrial | | Authors: | Khamrui, S, Lazarus, M.B. | | Deposit date: | 2022-08-25 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Characterization and structure of the human lysine-2-oxoglutarate reductase domain, a novel therapeutic target for treatment of glutaric aciduria type 1.
Open Biology, 12, 2022
|
|
7DBN
 
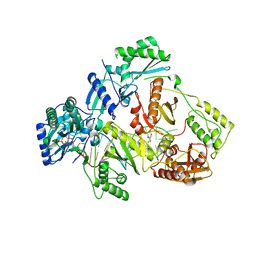 | | HIV-1 reverse transcriptase mutant Q151M/Y115F/F116Y/M184V/F160M:DNA:dCTP ternary complex | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA/RNA (38-MER), GLYCEROL, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2020-10-21 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Biochemical and Structural Properties of Entecavir-Resistant Hepatitis B Virus Polymerase with L180M/M204V Mutations.
J.Virol., 95, 2021
|
|
7DBM
 
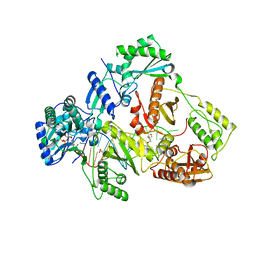 | | HIV-1 reverse transcriptase mutant Q151M/Y115F/F116Y/M184V:DNA:dGTP ternary complex | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA/RNA (38-MER), GLYCEROL, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2020-10-21 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Biochemical and Structural Properties of Entecavir-Resistant Hepatitis B Virus Polymerase with L180M/M204V Mutations.
J.Virol., 95, 2021
|
|
3AOX
 
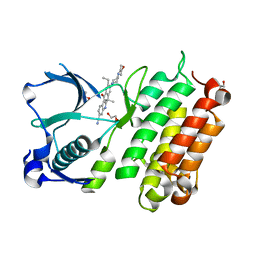 | | X-ray crystal structure of human anaplastic lymphoma kinase in complex with CH5424802 | | Descriptor: | 1,2-ETHANEDIOL, 9-ethyl-6,6-dimethyl-8-[4-(morpholin-4-yl)piperidin-1-yl]-11-oxo-6,11-dihydro-5H-benzo[b]carbazole-3-carbonitrile, ALK tyrosine kinase receptor | | Authors: | Nagel, S, Moertl, M, Jestel, A, Fukami, T.A. | | Deposit date: | 2010-10-08 | | Release date: | 2011-05-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | CH5424802, a selective ALK inhibitor capable of blocking the resistant gatekeeper mutant
Cancer Cell, 19, 2011
|
|
6IZY
 
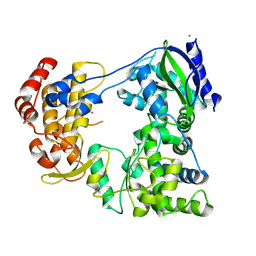 | |
7YFC
 
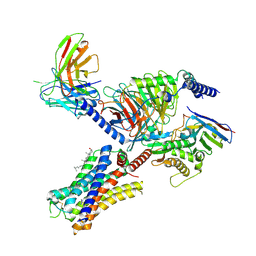 | | Cryo-EM structure of the histamine-bound histamine H4 receptor and Gq complex | | Descriptor: | CHOLESTEROL, Engineered G-alpha-q, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Im, D, Iwata, S, Asada, H. | | Deposit date: | 2022-07-08 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the agonists binding and receptor selectivity of human histamine H 4 receptor.
Nat Commun, 14, 2023
|
|
7YFD
 
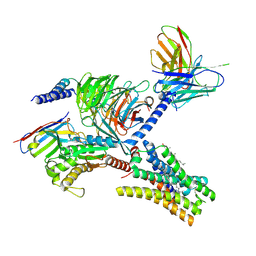 | | Cryo-EM structure of the imetit-bound histamine H4 receptor and Gq complex | | Descriptor: | 2-(1~{H}-imidazol-5-yl)ethyl carbamimidothioate, CHOLESTEROL, Engineered G-alpha-q, ... | | Authors: | Im, D, Iwata, S, Asada, H. | | Deposit date: | 2022-07-08 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the agonists binding and receptor selectivity of human histamine H 4 receptor.
Nat Commun, 14, 2023
|
|
7XVJ
 
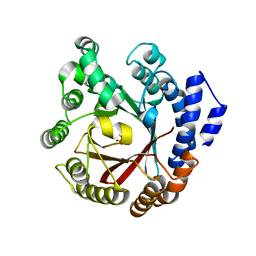 | | Crystal structure of CdpNPT in complex with harmol | | Descriptor: | 1-methyl-9~{H}-pyrido[3,4-b]indol-7-ol, Cyclic dipeptide N-prenyltransferase, PHOSPHATE ION | | Authors: | Nakashima, Y, Morita, H. | | Deposit date: | 2022-05-24 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Enzymatic formation of a prenyl beta-carboline by a fungal indole prenyltransferase.
J Nat Med, 76, 2022
|
|
6IZX
 
 | | The RNA-dependent RNA polymerase domain of dengue 2 NS5, bound with RK-0404678 | | Descriptor: | 2-oxo-2H-1,3-benzoxathiol-5-yl acetate, COBALT (II) ION, Genome polyprotein, ... | | Authors: | Shimizu, H, Sekine, S. | | Deposit date: | 2018-12-20 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discovery of a small molecule inhibitor targeting dengue virus NS5 RNA-dependent RNA polymerase.
Plos Negl Trop Dis, 13, 2019
|
|
2ZFU
 
 | | Structure of the methyltransferase-like domain of nucleomethylin | | Descriptor: | Cerebral protein 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Minami, H, Hashimoto, H, Murayama, A, Yanagisawa, J, Sato, M, Shimizu, T. | | Deposit date: | 2008-01-14 | | Release date: | 2008-12-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Epigenetic control of rDNA loci in response to intracellular energy status
Cell(Cambridge,Mass.), 133, 2008
|
|
6U3J
 
 | | Structure of the 2-oxoadipate dehydrogenase DHTKD1 | | Descriptor: | 2-oxoglutarate dehydrogenase E1 component DHKTD1, mitochondrial, MAGNESIUM ION, ... | | Authors: | Khamrui, S, Lazarus, M.B. | | Deposit date: | 2019-08-21 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Inhibition and Crystal Structure of the Human DHTKD1-Thiamin Diphosphate Complex.
Acs Chem.Biol., 15, 2020
|
|
3VVR
 
 | | Crystal structure of MATE in complex with MaD5 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Putative uncharacterized protein, macrocyclic peptide | | Authors: | Tanaka, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2012-07-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the drug extrusion mechanism by a MATE multidrug transporter.
Nature, 496, 2013
|
|
2RVD
 
 | |
3VVS
 
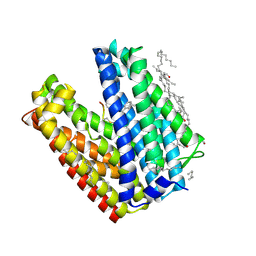 | | Crystal structure of MATE in complex with MaD3S | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Putative uncharacterized protein, macrocyclic peptide | | Authors: | Tanaka, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2012-07-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the drug extrusion mechanism by a MATE multidrug transporter.
Nature, 496, 2013
|
|
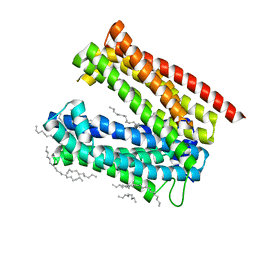
3VVO
 
 | |
3VVN
 
 | |
3VVP
 
 | | Crystal structure of MATE in complex with Br-NRF | | Descriptor: | 6-bromo-1-ethyl-4-oxo-7-(piperazin-1-yl)-1,4-dihydroquinoline-3-carboxylic acid, Putative uncharacterized protein | | Authors: | Tanaka, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2012-07-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structural basis for the drug extrusion mechanism by a MATE multidrug transporter.
Nature, 496, 2013
|
|
7FFH
 
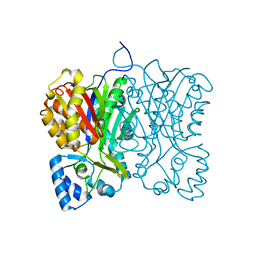 | | Diarylpentanoid-producing polyketide synthase (N199L mutant) | | Descriptor: | Type III polyketide synthase | | Authors: | Morita, H, Wong, C.P, Liu, Q, Takeshi, K, Lee, Y, Nakashima, Y. | | Deposit date: | 2021-07-23 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of a diarylpentanoid-producing polyketide synthase revealing an unusual biosynthetic pathway of 2-(2-phenylethyl)chromones in agarwood.
Nat Commun, 13, 2022
|
|
3WBN
 
 | | Crystal structure of MATE in complex with MaL6 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, MaL6, Putative uncharacterized protein | | Authors: | Tanaka, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2013-05-20 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for the drug extrusion mechanism by a MATE multidrug transporter.
Nature, 496, 2013
|
|
3W4T
 
 | | Crystal structure of MATE P26A mutant | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Putative uncharacterized protein | | Authors: | Tanaka, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2013-01-16 | | Release date: | 2013-04-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Structural basis for the drug extrusion mechanism by a MATE multidrug transporter.
Nature, 496, 2013
|
|
