6GI6
 
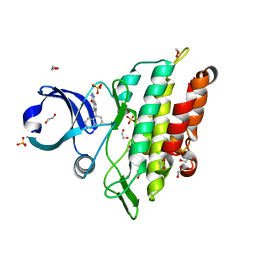 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with a Quinazolinone based ALK2 inhibitor with a 5-methyl core. | | Descriptor: | 1,2-ETHANEDIOL, 5-methyl-6-quinolin-5-yl-3~{H}-quinazolin-4-one, Activin receptor type-1, ... | | Authors: | Williams, E, Hudson, L, Bezerra, G.A, Sorrell, F, Mahajan, P, Kupinska, K, Hoelder, S, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2018-05-10 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Novel Quinazolinone Inhibitors of ALK2 Flip between Alternate Binding Modes: Structure-Activity Relationship, Structural Characterization, Kinase Profiling, and Cellular Proof of Concept.
J. Med. Chem., 61, 2018
|
|
6GIP
 
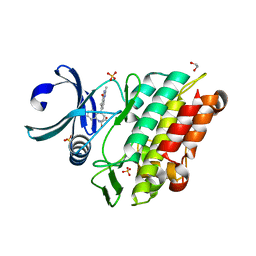 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with a Quinazolinone based ALK2 inhibitor with a 2, 5-dimethyl core. | | Descriptor: | 1,2-ETHANEDIOL, 2,5-dimethyl-6-quinolin-4-yl-3~{H}-quinazolin-4-one, Activin receptor type-1, ... | | Authors: | Williams, E, Hudson, L, Bezerra, G.A, Sorrell, F, Mathea, S, Chen, Z, Mahajan, P, Kupinska, K, Hoelder, S, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2018-05-14 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Novel Quinazolinone Inhibitors of ALK2 Flip between Alternate Binding Modes: Structure-Activity Relationship, Structural Characterization, Kinase Profiling, and Cellular Proof of Concept.
J. Med. Chem., 61, 2018
|
|
6C1Y
 
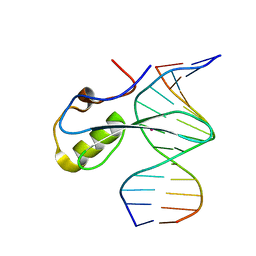 | | mbd of human mecp2 in complex with methylated DNA | | Descriptor: | 12-mer DNA, Methyl-CpG-binding protein 2, UNKNOWN ATOM OR ION | | Authors: | Liu, K, Bian, C, Tempel, W, Wernimont, A.K, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-01-05 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Plasticity at the DNA recognition site of the MeCP2 mCG-binding domain.
Biochim Biophys Acta Gene Regul Mech, 1862, 2019
|
|
4WSQ
 
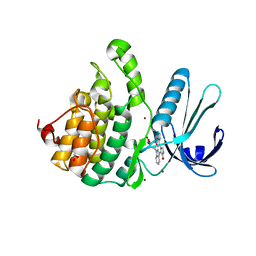 | | Crystal Structure of Adaptor Protein 2 Associated Kinase (AAK1) in complex with small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, AP2-associated protein kinase 1, K-252A, ... | | Authors: | Sorrell, F.J, Elkins, J.M, Krojer, T, Williams, E, Abdul, K, Gileadi, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-10-28 | | Release date: | 2014-11-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Family-wide Structural Analysis of Human Numb-Associated Protein Kinases.
Structure, 24, 2016
|
|
4RCJ
 
 | | Crystal structure of YTHDF1 YTH domain in complex with 5mer m6A RNA | | Descriptor: | RNA (5'-R(*GP*GP*(6MZ)P*CP*U)-3'), UNKNOWN ATOM OR ION, YTH domain-containing family protein 1 | | Authors: | Tempel, W, Xu, C, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-09-16 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for the Discriminative Recognition of N6-Methyladenosine RNA by the Human YT521-B Homology Domain Family of Proteins.
J.Biol.Chem., 290, 2015
|
|
7ZKF
 
 | | Crystal Structure of human Brachyury G177D variant in complex with CF-5-86 | | Descriptor: | N-(4-((2-fluorophenyl)(hydroxy)methyl)phenyl)acetamide, PHOSPHATE ION, T-box transcription factor T | | Authors: | Newman, J.A, Gavard, A, Aitkenhead, H, Imprachim, N, Sherestha, L, Burgess-Brown, N.A, von Delft, F, Bountra, C, Gileadi, O. | | Deposit date: | 2022-04-12 | | Release date: | 2022-06-22 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural insights into human brachyury DNA recognition and discovery of progressible binders for cancer therapy.
Nat Commun, 16, 2025
|
|
6GIN
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with an Quinazolinone based ALK2 inhibitor with a 4-morpholinophenyl solvent accessible group. | | Descriptor: | 1,2-ETHANEDIOL, 3-(4-morpholin-4-ylphenyl)-6-quinolin-4-yl-quinazolin-4-one, Activin receptor type-1, ... | | Authors: | Williams, E, Hudson, L, Bezerra, G.A, Kopec, J, Mahajan, P, Kupinska, K, Hoelder, S, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2018-05-14 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel Quinazolinone Inhibitors of ALK2 Flip between Alternate Binding Modes: Structure-Activity Relationship, Structural Characterization, Kinase Profiling, and Cellular Proof of Concept.
J. Med. Chem., 61, 2018
|
|
6EK6
 
 | | Crystal structure of KDM5B in complex with S49195a. | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Lysine-specific demethylase 5B,Lysine-specific demethylase 5B, ... | | Authors: | Srikannathasan, V, Szykowska, A, Newman, J.A, Ruda, G.F, Strain-Damerell, C, Burgess-Brown, N.A, Vazquez-Rodriguez, S, Wright, M, Brennan, P.E, Arrowsmith, C.H, Edwards, A, Bountra, C, Oppermann, U, Huber, K, von Delft, F. | | Deposit date: | 2017-09-25 | | Release date: | 2018-03-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of KDM5B in complex with S49195a.
To be published
|
|
4YU2
 
 | | Crystal structure of DYRK1A with harmine-derivatized AnnH-75 inhibitor | | Descriptor: | (1-chloro-7-methoxy-9H-beta-carbolin-9-yl)acetonitrile, Dual specificity tyrosine-phosphorylation-regulated kinase 1A, SULFATE ION, ... | | Authors: | Chaikuad, A, Wurzlbauer, A, Nowak, R, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bracher, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-03-18 | | Release date: | 2015-03-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | How to Separate Kinase Inhibition from Undesired Monoamine Oxidase A Inhibition-The Development of the DYRK1A Inhibitor AnnH75 from the Alkaloid Harmine.
Molecules, 25, 2020
|
|
4Z31
 
 | | Crystal structure of the RC3H2 ROQ domain in complex with stem-loop and double-stranded forms of RNA | | Descriptor: | CHLORIDE ION, RNA (5'-R(*A)-D(P*UP*GP*UP*UP*CP*UP*GP*UP*GP*AP*AP*CP*AP*C)-3'), Roquin-2, ... | | Authors: | DONG, A, ZHANG, Q, TEMPEL, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, TONG, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-03-30 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | New Insights into the RNA-Binding and E3 Ubiquitin Ligase Activities of Roquins.
Sci Rep, 5, 2015
|
|
6EIN
 
 | | Crystal structure of KDM5B in complex with S49365a. | | Descriptor: | 1,2-ETHANEDIOL, 8-[4-[2-(4-propanoylpiperazin-1-yl)ethyl]pyrazol-1-yl]-3~{H}-pyrido[3,4-d]pyrimidin-4-one, Lysine-specific demethylase 5B,Lysine-specific demethylase 5B, ... | | Authors: | Srikannathasan, V, Newman, J.A, Szykowska, A, Wright, M, Ruda, G.F, Vazquez-Rodriguez, S.A, Kupinska, K, Strain-Damerell, C, Burgess-Brown, N.A, Arrowsmith, C.H, Edwards, A, Bountra, C, Oppermann, U, Huber, K, von Delft, F. | | Deposit date: | 2017-09-19 | | Release date: | 2018-05-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of KDM5B in complex with S49365a.
To Be Published
|
|
6EIM
 
 | | Human STK10 bound to GW683134A | | Descriptor: | 1,2-ETHANEDIOL, Serine/threonine-protein kinase 10, ~{N}-[5-[4-[[2-fluoranyl-5-(trifluoromethyl)phenyl]carbamoylamino]phenoxy]-1~{H}-benzimidazol-2-yl]furan-2-carboxamide | | Authors: | Sorrell, F.J, Berger, B.-T, Salah, E, von Delft, F, Bountra, C, Arrowsmith, C, Edwards, A.M, Knapp, S, Elkins, J.M. | | Deposit date: | 2017-09-19 | | Release date: | 2017-11-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Human STK10 bound to GW683134
To Be Published
|
|
6EJ1
 
 | | Crystal structure of KDM5B in complex with KDOPZ48a. | | Descriptor: | 1,2-ETHANEDIOL, 5-[1-[1-(2-chloranylethanoyl)piperidin-4-yl]pyrazol-4-yl]-7-oxidanylidene-6-propan-2-yl-4~{H}-pyrazolo[1,5-a]pyrimidine-3-carbonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Srikannathasan, V, Newman, J.A, Szykowska, A, Wright, M, Ruda, G.F, Vazquez-Rodriguez, S.A, Kupinska, K, Strain-Damerell, C, Burgess-Brown, N.A, Arrowsmith, C.H, Edwards, A, Bountra, C, Oppermann, U, Huber, K, von Delft, F. | | Deposit date: | 2017-09-19 | | Release date: | 2018-05-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of KDM5B in complex with KDOPZ48a.
to be published
|
|
3UWP
 
 | | Crystal structure of Dot1l in complex with 5-iodotubercidin | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Yu, W, Tempel, W, Smil, D, Schapira, M, Li, Y, Vedadi, M, Nguyen, K.T, Wernimont, A.K, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-12-02 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Catalytic site remodelling of the DOT1L methyltransferase by selective inhibitors.
Nat Commun, 3, 2012
|
|
8A7N
 
 | | Crystal Structure of human Brachyury G177D variant in complex with (S)-N-(3-aminopropyl)-3-((1-(2-fluorophenyl)-2-oxopyrrolidin-3-yl)amino)-N-methylbenzamide (CF-2-125) | | Descriptor: | N-(3-azanylpropyl)-3-[[(3S)-1-(2-fluorophenyl)-2-oxidanylidene-pyrrolidin-3-yl]amino]-N-methyl-benzamide, PHOSPHATE ION, T-box transcription factor T | | Authors: | Newman, J.A, Gavard, A, Aitkenhead, H, Imprachim, N, Sherestha, L, Burgess-Brown, N.A, von Delft, F, Bountra, C, Gileadi, O. | | Deposit date: | 2022-06-21 | | Release date: | 2022-10-05 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into human brachyury DNA recognition and discovery of progressible binders for cancer therapy.
Nat Commun, 16, 2025
|
|
4RCI
 
 | | Crystal structure of YTHDF1 YTH domain | | Descriptor: | UNKNOWN ATOM OR ION, YTH domain-containing family protein 1 | | Authors: | Xu, C, Tempel, W, Liu, K, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-09-16 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Basis for the Discriminative Recognition of N6-Methyladenosine RNA by the Human YT521-B Homology Domain Family of Proteins.
J.Biol.Chem., 290, 2015
|
|
4QC3
 
 | | Crystal structure of human BAZ2B bromodomain in complex with a diacetylated histone 4 peptide (H4K8acK12ac) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B, diacetylated histone 4 peptide (H4K8acK12ac) | | Authors: | Tallant, C, Jose, B, Picaud, S, Chaikuad, A, Filippakopoulos, P, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-05-09 | | Release date: | 2014-05-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis of histone tail recognition by human TIP5 PHD finger and bromodomain of the chromatin remodeling complex NoRC.
Structure, 23, 2015
|
|
4QC1
 
 | | Crystal structure of human BAZ2B bromodomain in complex with an acetylated histone 3 peptide (H3K14ac) | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2B, SULFATE ION, ZINC ION, ... | | Authors: | Tallant, C, Jose, B, Picaud, S, Chaikuad, A, Filippakopoulos, P, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-05-09 | | Release date: | 2014-05-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Molecular basis of histone tail recognition by human TIP5 PHD finger and bromodomain of the chromatin remodeling complex NoRC.
Structure, 23, 2015
|
|
3U67
 
 | | Crystal structure of the N-terminal domain of Hsp90 from Leishmania major(LmjF33.0312)in complex with ADP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, Heat shock protein 83-1, ... | | Authors: | Pizarro, J.C, Wernimont, A.K, Hutchinson, A, Mackenzie, F, Fairlamb, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Ferguson, M.A.J, Hui, R, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-10-12 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Exploring the Trypanosoma brucei Hsp83 potential as a target for structure guided drug design.
PLoS Negl Trop Dis, 7, 2013
|
|
3DB3
 
 | | Crystal structure of the tandem tudor domains of the E3 ubiquitin-protein ligase UHRF1 in complex with trimethylated histone H3-K9 peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Trimethylated histone H3-K9 peptide | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Dong, A, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-30 | | Release date: | 2008-09-16 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Recognition of multivalent histone states associated with heterochromatin by UHRF1 protein.
J.Biol.Chem., 286, 2011
|
|
4PZI
 
 | | Zinc finger region of MLL2 in complex with CpG DNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*CP*CP*GP*GP*TP*GP*GP*C)-3'), Histone-lysine N-methyltransferase 2B, UNKNOWN ATOM OR ION, ... | | Authors: | Chao, X, Tempel, W, Liu, K, Dong, A, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-03-31 | | Release date: | 2014-06-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | DNA Sequence Recognition of Human CXXC Domains and Their Structural Determinants.
Structure, 26, 2018
|
|
3E0C
 
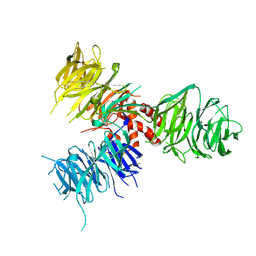 | | Crystal Structure of DNA Damage-Binding protein 1(DDB1) | | Descriptor: | DNA damage-binding protein 1 | | Authors: | Amaya, M.F, Xu, L, Hao, H, Bountra, C, Wickstroem, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-07-31 | | Release date: | 2008-09-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structure and function of WD40 domain proteins.
Protein Cell, 2, 2011
|
|
3EPZ
 
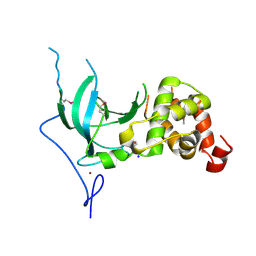 | | Structure of the replication foci-targeting sequence of human DNA cytosine methyltransferase DNMT1 | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, GLYCEROL, SODIUM ION, ... | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-09-30 | | Release date: | 2008-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The replication focus targeting sequence (RFTS) domain is a DNA-competitive inhibitor of Dnmt1.
J.Biol.Chem., 286, 2011
|
|
3EAE
 
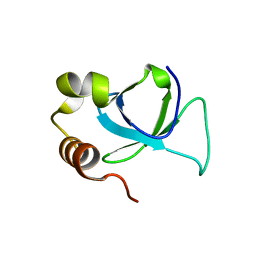 | | PWWP domain of human hepatoma-derived growth factor 2 (HDGF2) | | Descriptor: | Hepatoma-derived growth factor-related protein 2 | | Authors: | Amaya, M.F, Zeng, H, Mackenzie, F, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-08-25 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural and Histone Binding Ability Characterizations of Human PWWP Domains.
Plos One, 6, 2011
|
|
4OGJ
 
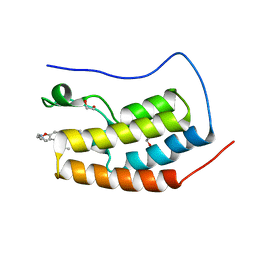 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor TG-101348 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N-tert-butyl-3-{[5-methyl-2-({4-[2-(pyrrolidin-1-yl)ethoxy]phenyl}amino)pyrimidin-4-yl]amino}benzenesulfonamide | | Authors: | Filippakopoulos, P, Picaud, S, Jose, B, Martin, S, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-01-16 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Dual kinase-bromodomain inhibitors for rationally designed polypharmacology.
Nat.Chem.Biol., 10, 2014
|
|
