4GOV
 
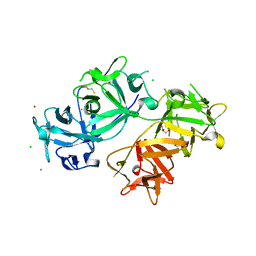 | | The crystal structure of human fascin 1 S39D mutant | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Yang, S.Y, Huang, F.K, Huang, J, Chen, S, Jakoncic, J, Leo-Macias, A, Diaz-Avalos, R, Chen, L, Zhang, J.J, Huang, X.Y. | | Deposit date: | 2012-08-20 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular mechanism of fascin function in filopodial formation.
J.Biol.Chem., 288, 2013
|
|
4GP0
 
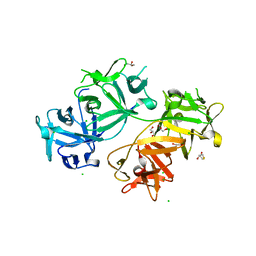 | | The crystal structure of human fascin 1 R149A K150A R151A mutant | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BROMIDE ION, ... | | Authors: | Yang, S.Y, Huang, F.K, Huang, J, Chen, S, Jakoncic, J, Leo-Macias, A, Diaz-Avalos, R, Chen, L, Zhang, J.J, Huang, X.Y. | | Deposit date: | 2012-08-20 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism of fascin function in filopodial formation.
J.Biol.Chem., 288, 2013
|
|
4GOY
 
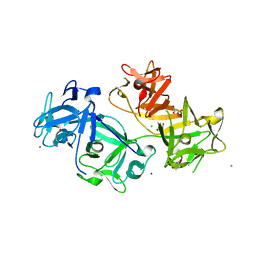 | | The crystal structure of human fascin 1 K41A mutant | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Yang, S.Y, Huang, F.K, Huang, J, Chen, S, Jakoncic, J, Leo-Macias, A, Diaz-Avalos, R, Chen, L, Zhang, J.J, Huang, X.Y. | | Deposit date: | 2012-08-20 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular mechanism of fascin function in filopodial formation.
J.Biol.Chem., 288, 2013
|
|
1OZ9
 
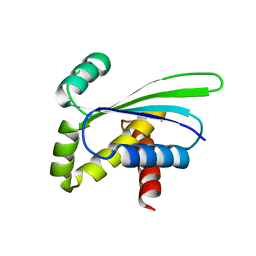 | | Crystal structure of AQ_1354, a hypothetical protein from Aquifex aeolicus | | Descriptor: | Hypothetical protein AQ_1354 | | Authors: | Oganesyan, V, Busso, D, Brandsen, J, Chen, S, Jancarik, J, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2003-04-08 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.894 Å) | | Cite: | Structure of the hypothetical protein AQ_1354 from Aquifex aeolicus.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
4GHQ
 
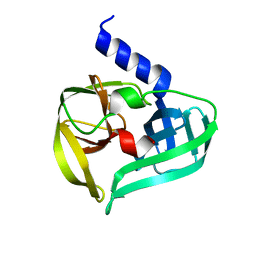 | | Crystal structure of EV71 3C proteinase | | Descriptor: | 3C proteinase | | Authors: | Chen, C, Wu, C, Cai, Q, Li, N, Peng, X, Cai, Y, Yin, K, Chen, X, Wang, X, Zhang, R, Liu, L, Chen, S, Li, J, Lin, T. | | Deposit date: | 2012-08-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of Enterovirus 71 3C proteinase (strain E2004104-TW-CDC) and its complex with rupintrivir
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6ITY
 
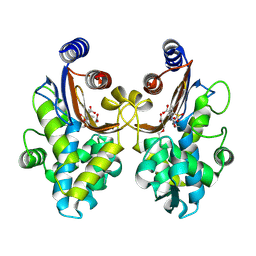 | | CTX-M-64 sulbactam complex | | Descriptor: | ACRYLIC ACID, Beta-lactamase, TRANS-ENAMINE INTERMEDIATE OF SULBACTAM | | Authors: | Cheng, Q, Chen, S. | | Deposit date: | 2018-11-26 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural Insight into the Mechanism of Inhibitor Resistance in CTX-M-199, a CTX-M-64 Variant Carrying the S130T Substitution.
Acs Infect Dis., 6, 2020
|
|
6J2B
 
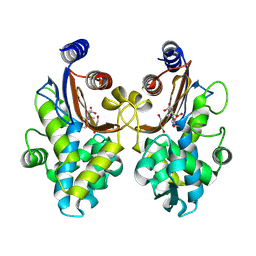 | | CTX-M-64 beta-lactamase S130T sulbactam complex | | Descriptor: | Beta-lactamase, GLYCEROL, TRANS-ENAMINE INTERMEDIATE OF SULBACTAM | | Authors: | Cheng, Q, Chen, S. | | Deposit date: | 2018-12-31 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structural Insight into the Mechanism of Inhibitor Resistance in CTX-M-199, a CTX-M-64 Variant Carrying the S130T Substitution.
Acs Infect Dis., 6, 2020
|
|
6J2O
 
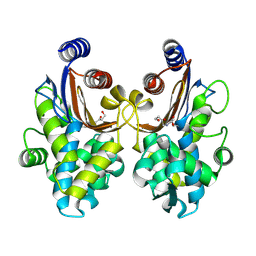 | | Crystal structure of CTX-M-64 clavulanic acid complex | | Descriptor: | (2E)-3-[(4-hydroxy-2-oxobutyl)amino]prop-2-enal, Beta-lactamase | | Authors: | Cheng, Q, Chen, S. | | Deposit date: | 2019-01-02 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insight into the Mechanism of Inhibitor Resistance in CTX-M-199, a CTX-M-64 Variant Carrying the S130T Substitution.
Acs Infect Dis., 6, 2020
|
|
6J25
 
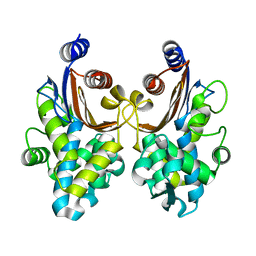 | | CTX-M-64 beta-lactamase mutant-S130T | | Descriptor: | Beta-lactamase | | Authors: | Cheng, Q, Chen, S. | | Deposit date: | 2018-12-30 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Insight into the Mechanism of Inhibitor Resistance in CTX-M-199, a CTX-M-64 Variant Carrying the S130T Substitution.
Acs Infect Dis., 6, 2020
|
|
6J2K
 
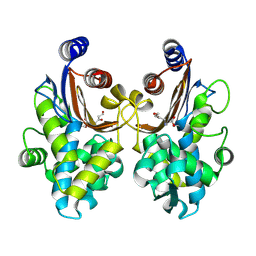 | | CTX-M-64 beta-lactamase S130T clavulanic acid complex | | Descriptor: | (2E)-3-[(4-hydroxy-2-oxobutyl)amino]prop-2-enal, Beta-lactamase | | Authors: | Cheng, Q, Chen, S. | | Deposit date: | 2019-01-01 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structural Insight into the Mechanism of Inhibitor Resistance in CTX-M-199, a CTX-M-64 Variant Carrying the S130T Substitution.
Acs Infect Dis., 6, 2020
|
|
4KQW
 
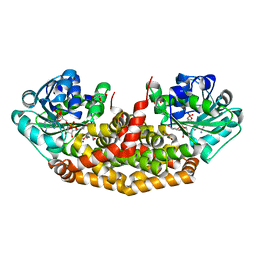 | | The structure of the Slackia exigua KARI in complex with NADP | | Descriptor: | Ketol-acid reductoisomerase, L(+)-TARTARIC ACID, MAGNESIUM ION, ... | | Authors: | Brinkmann-Chen, S, Flock, T, Cahn, J.K.B, Snow, C.D, Brustad, E.M, Mcintosh, J.A, Meinhold, P, Zhang, L, Arnold, F.H. | | Deposit date: | 2013-05-15 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | General approach to reversing ketol-acid reductoisomerase cofactor dependence from NADPH to NADH.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6JV4
 
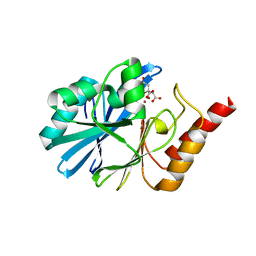 | | Crystal structure of metallo-beta-lactamase VMB-1 | | Descriptor: | CITRIC ACID, VMB-1, ZINC ION | | Authors: | Cheng, Q, Chen, S. | | Deposit date: | 2019-04-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Genetic and Biochemical Characterization of VMB-1, a Novel Metallo-beta-Lactamase Encoded by a Conjugative, Broad-Host Range IncC Plasmid from Vibrio spp.
Adv Biosyst, 4, 2020
|
|
4KQX
 
 | | Mutant Slackia exigua KARI DDV in complex with NAD and an inhibitor | | Descriptor: | HISTIDINE, Ketol-acid reductoisomerase, MAGNESIUM ION, ... | | Authors: | Brinkmann-Chen, S, Flock, T, Cahn, J.K.B, Snow, C.D, Brustad, E.M, Mcintosh, J.A, Meinhold, P, Zhang, L, Arnold, F.H. | | Deposit date: | 2013-05-15 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | General approach to reversing ketol-acid reductoisomerase cofactor dependence from NADPH to NADH.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6WIV
 
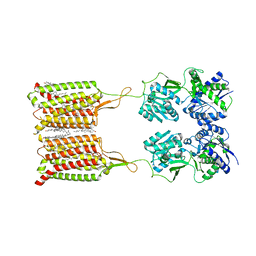 | | Structure of human GABA(B) receptor in an inactive state | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-{[(9Z)-octadec-9-enoyl]oxy}propyl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Park, J, Fu, Z, Frangaj, A, Liu, J, Mosyak, L, Shen, T, Slavkovich, V.N, Ray, K.M, Taura, J, Cao, B, Geng, Y, Zuo, H, Kou, Y, Grassucci, R, Chen, S, Liu, Z, Lin, X, Williams, J.P, Rice, W.J, Eng, E.T, Huang, R.K, Soni, R.K, Kloss, B, Yu, Z, Javitch, J.A, Hendrickson, W.A, Slesinger, P.A, Quick, M, Graziano, J, Yu, H, Fiehn, O, Clarke, O.B, Frank, J, Fan, Q.R. | | Deposit date: | 2020-04-10 | | Release date: | 2020-07-01 | | Last modified: | 2020-08-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of human GABABreceptor in an inactive state.
Nature, 584, 2020
|
|
1H69
 
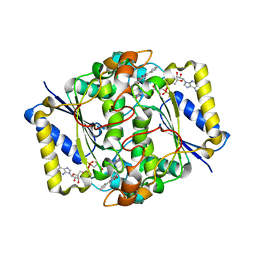 | | CRYSTAL STRUCTURE OF HUMAN NAD[P]H-QUINONE OXIDOREDUCTASE CO WITH 2,3,5,6,TETRAMETHYL-P-BENZOQUINONE (DUROQUINONE) AT 2.5 ANGSTROM RESOLUTION | | Descriptor: | 3-(HYDROXYMETHYL)-1-METHYL-5-(2-METHYLAZIRIDIN-1-YL)-2-PHENYL-1H-INDOLE-4,7-DIONE, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H DEHYDROGENASE [QUINONE] 1 | | Authors: | Faig, M, Bianchet, M.A, Chen, S, Winski, S, Ross, D, Amzel, L.M. | | Deposit date: | 2001-06-08 | | Release date: | 2001-09-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure-Based Development of Anticancer Drugs: Complexes of Nad(P)H:Quinone Oxidoreductase 1 with Chemotherapeutic Quinones
Structure, 9, 2001
|
|
4I5X
 
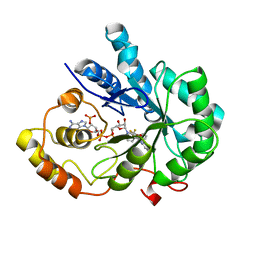 | | Crystal Structure Of AKR1B10 Complexed With NADP+ And Flufenamic acid | | Descriptor: | 2-[[3-(TRIFLUOROMETHYL)PHENYL]AMINO] BENZOIC ACID, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, L, Zheng, X, Chen, S, Zhai, J, Zhang, H, Zhao, Y. | | Deposit date: | 2012-11-29 | | Release date: | 2013-10-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibitor selectivity between aldo-keto reductase superfamily members AKR1B10 and AKR1B1: Role of Trp112 (Trp111)
Febs Lett., 587, 2013
|
|
4GQG
 
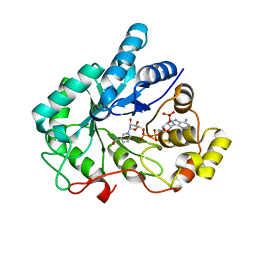 | | Crystal structure of AKR1B10 complexed with NADP+ | | Descriptor: | Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, L, Zheng, X, Chen, S, Zhai, J, Hu, X. | | Deposit date: | 2012-08-23 | | Release date: | 2013-08-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Inhibitor selectivity between aldo-keto reductase superfamily members AKR1B10 and AKR1B1: Role of Trp112 (Trp111)
Febs Lett., 587, 2013
|
|
5ZB7
 
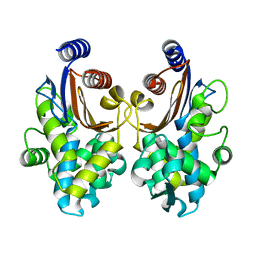 | | CTX-M-64 apoenzyme | | Descriptor: | Beta-lactamase | | Authors: | Cheng, Q, Chen, S. | | Deposit date: | 2018-02-10 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural Insight into the Mechanism of Inhibitor Resistance in CTX-M-199, a CTX-M-64 Variant Carrying the S130T Substitution.
Acs Infect Dis., 6, 2020
|
|
5A8H
 
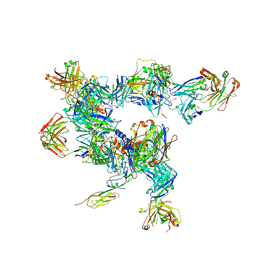 | | cryo-ET subtomogram averaging of BG505 SOSIP.664 in complex with sCD4, 17b, and 8ANC195 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB OF BROADLY NEUTRALIZING ANTIBODY 17B, FAB OF BROADLY NEUTRALIZING ANTIBODY 8ANC195 VARIANT G52K5, ... | | Authors: | Scharf, L, Wang, H, Gao, H, Chen, S, McDowall, A, Bjorkman, P. | | Deposit date: | 2015-07-15 | | Release date: | 2015-08-05 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | Broadly Neutralizing Antibody 8ANC195 Recognizes Closed and Open States of HIV-1 Env.
Cell, 162, 2015
|
|
5A7X
 
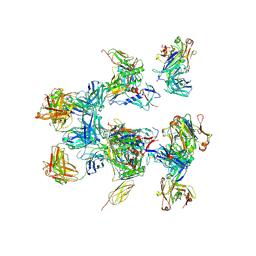 | | negative stain EM of BG505 SOSIP.664 in complex with sCD4, 17b, and 8ANC195 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB OF BROADLY NEUTRALIZING ANTIBODY 17B, FAB OF BROADLY NEUTRALIZING ANTIBODY 8ANC195, ... | | Authors: | Scharf, L, Wang, H, Gao, H, Chen, S, McDowall, A, Bjorkman, P. | | Deposit date: | 2015-07-10 | | Release date: | 2015-08-05 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Broadly Neutralizing Antibody 8Anc195 Recognizes Closed and Open States of HIV-1 Env.
Cell(Cambridge,Mass.), 162, 2015
|
|
8JMP
 
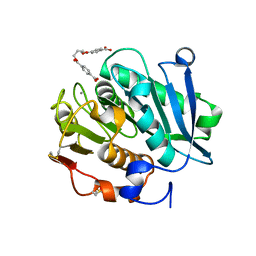 | | Structure of a leaf-branch compost cutinase, ICCG in complex with 1,4-butanediol terephthalate | | Descriptor: | 4-[4-(4-carboxyphenyl)carbonyloxybutoxycarbonyl]benzoic acid, CALCIUM ION, Leaf-branch compost cutinase | | Authors: | Yang, Y, Xue, T, Zheng, Y, Cheng, S, Guo, R.-T, Chen, C.-C. | | Deposit date: | 2023-06-05 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Remodeling the polymer-binding cavity to improve the efficacy of PBAT-degrading enzyme.
J Hazard Mater, 464, 2023
|
|
8JMO
 
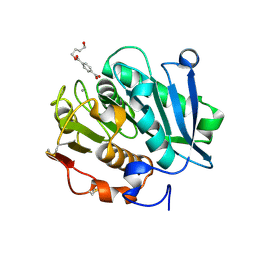 | | Structure of a leaf-branch compost cutinase, ICCG in complex with 4-((4-Hydroxybutoxy)carbonyl)benzoic acid | | Descriptor: | 4-(4-oxidanylbutoxycarbonyl)benzoic acid, CALCIUM ION, Leaf-branch compost cutinase | | Authors: | Yang, Y, Xue, T, Zheng, Y, Cheng, S, Guo, R.-T, Chen, C.-C. | | Deposit date: | 2023-06-05 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Remodeling the polymer-binding cavity to improve the efficacy of PBAT-degrading enzyme.
J Hazard Mater, 464, 2023
|
|
5FNA
 
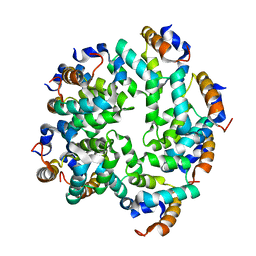 | | Cryo-EM reconstruction of caspase-1 CARD | | Descriptor: | Caspase-1 | | Authors: | Li, Y, Lu, A, Schmidt, F.I, Yin, Q, Chen, S, Fu, T.M, Tong, A.B, Ploegh, H.L, Mao, Y, Wu, H. | | Deposit date: | 2015-11-11 | | Release date: | 2016-03-30 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Molecular Basis of Caspase-1 Polymerization and its Inhibition by a Novel Capping Mechanism
Nat.Struct.Mol.Biol., 23, 2016
|
|
2H53
 
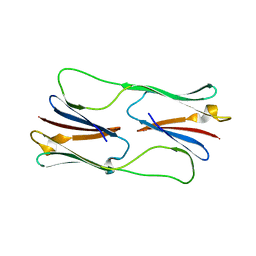 | | Multiple distinct assemblies reveal conformational flexibility in the small heat shock protein Hsp26 | | Descriptor: | small heat shock protein Hsp26 | | Authors: | White, H.E, Orlova, E.V, Chen, S, Wang, L, Ignatiou, A, Gowen, B, Stromer, T, Franzmann, T.M, Haslbeck, M, Buchner, J, Saibil, H.R. | | Deposit date: | 2006-05-25 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Multiple distinct assemblies reveal conformational flexibility in the small heat shock protein hsp26
Structure, 14, 2006
|
|
2H50
 
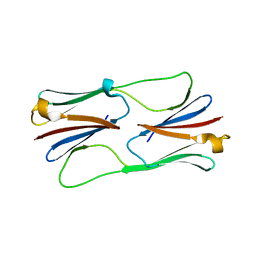 | | Multiple distinct assemblies reveal conformational flexibility in the small heat shock protein Hsp26 | | Descriptor: | small heat shock protein Hsp26 | | Authors: | White, H.E, Orlova, E.V, Chen, S, Wang, L, Ignatiou, A, Gowen, B, Stromer, T, Franzmann, T.M, Haslbeck, M, Buchner, J, Saibil, H.R. | | Deposit date: | 2006-05-25 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (10.8 Å) | | Cite: | Multiple distinct assemblies reveal conformational flexibility in the small heat shock protein hsp26
Structure, 14, 2006
|
|
