6E67
 
 | | Structure of beta2 adrenergic receptor fused to a Gs peptide | | Descriptor: | 8-[(1R)-2-{[1,1-dimethyl-2-(2-methylphenyl)ethyl]amino}-1-hydroxyethyl]-5-hydroxy-2H-1,4-benzoxazin-3(4H)-one, Beta-2 adrenergic receptor,Endolysin,Guanine nucleotide-binding protein G(s) subunit alpha isoforms short,Beta-2 adrenergic receptor chimera | | Authors: | Liu, X, Xu, X, Hilger, D, Tiemann, J, Liu, H, Du, Y, Hirata, K, Sun, X, Guixa-Gonzalez, R, Mathiesen, J, Hildebrand, P, Kobilka, B. | | Deposit date: | 2018-07-24 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural Insights into the Process of GPCR-G Protein Complex Formation.
Cell, 177, 2019
|
|
7MYN
 
 | | Cryo-EM Structure of p110alpha in complex with p85alpha | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Liu, X, Yang, S, Hart, J.R, Xu, Y, Zou, X, Zhang, H, Zhou, Q, Xia, T, Zhang, Y, Yang, D, Wang, M.-W, Vogt, P.K. | | Deposit date: | 2021-05-21 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Cryo-EM structures of PI3K alpha reveal conformational changes during inhibition and activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MYO
 
 | | Cryo-EM structure of p110alpha in complex with p85alpha inhibited by BYL-719 | | Descriptor: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Liu, X, Yang, S, Hart, J.R, Xu, Y, Zou, X, Zhang, H, Zhou, Q, Xia, T, Zhang, Y, Yang, D, Wang, M.-W, Vogt, P.K. | | Deposit date: | 2021-05-21 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Cryo-EM structures of PI3K alpha reveal conformational changes during inhibition and activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5MV2
 
 | | Crystal structure of the E protein of the Japanese encephalitis live attenuated vaccine virus | | Descriptor: | E protein | | Authors: | Liu, X, Zhao, X, Na, R, Li, L, Warkentin, E, Witt, J, Lu, X, Wei, Y, Peng, G, Li, Y, Wang, J. | | Deposit date: | 2017-01-14 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure differences of Japanese encephalitis virus SA14 and SA14-14-2 E proteins elucidate the virulence attenuation mechanism.
Protein Cell, 10, 2019
|
|
5MV1
 
 | | Crystal structure of the E protein of the Japanese encephalitis virulent virus | | Descriptor: | E protein | | Authors: | Liu, X, Zhao, X, Na, R, Li, L, Warkentin, E, Witt, J, Lu, X, Wei, Y, Peng, G, Li, Y, Wang, J. | | Deposit date: | 2017-01-14 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The structure differences of Japanese encephalitis virus SA14 and SA14-14-2 E proteins elucidate the virulence attenuation mechanism.
Protein Cell, 10, 2019
|
|
6X6E
 
 | |
6X6D
 
 | |
3CI9
 
 | | Crystal Structure of the human HSBP1 | | Descriptor: | Heat shock factor-binding protein 1 | | Authors: | Liu, X, Xu, L, Liu, Y, Zhu, G, Zhang, X.C, Li, X, Rao, Z. | | Deposit date: | 2008-03-11 | | Release date: | 2009-01-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the hexamer of human heat shock factor binding protein 1
Proteins, 75, 2009
|
|
5HRA
 
 | | Crystal structure of an aspartate/glutamate racemase in complex with D-aspartate | | Descriptor: | D-ASPARTIC ACID, aspartate/glutamate racemase | | Authors: | Liu, X, Gao, F, Ma, Y, Liu, S, Cui, Y, Yuan, Z, Kang, X. | | Deposit date: | 2016-01-23 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.597 Å) | | Cite: | Crystal structure and molecular mechanism of an aspartate/glutamate racemase from Escherichia coli O157
Febs Lett., 590, 2016
|
|
6EXV
 
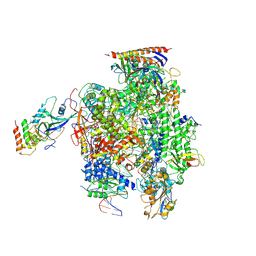 | | Structure of mammalian RNA polymerase II elongation complex inhibited by Alpha-amanitin | | Descriptor: | AMATOXIN, DNA (25-MER), DNA (36-MER), ... | | Authors: | Liu, X, Farnung, L, Wigge, C, Cramer, P. | | Deposit date: | 2017-11-09 | | Release date: | 2018-03-21 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of a mammalian RNA polymerase II elongation complex inhibited by alpha-amanitin.
J. Biol. Chem., 293, 2018
|
|
1LNU
 
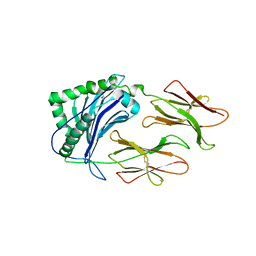 | | CRYSTAL STRUCTURE OF CLASS II MHC MOLECULE IAb BOUND TO EALPHA3K PEPTIDE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, H-2 class II histocompatibility antigen, A beta chain, ... | | Authors: | Liu, X, Dai, S, Crawford, F, Fruge, R, Marrack, P, Kappler, J. | | Deposit date: | 2002-05-03 | | Release date: | 2002-08-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Alternate interactions define the binding of peptides to the MHC molecule IA(b).
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
5HRC
 
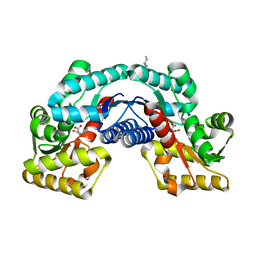 | | Crystal structure of an aspartate/glutamate racemase in complex with L-aspartate | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ASPARTIC ACID, aspartate/glutamate racemase | | Authors: | Liu, X, Gao, F, Ma, Y, Liu, S, Cui, Y, Yuan, Z, Kang, X. | | Deposit date: | 2016-01-23 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.765 Å) | | Cite: | Crystal structure and molecular mechanism of an aspartate/glutamate racemase from Escherichia coli O157
Febs Lett., 590, 2016
|
|
2B9D
 
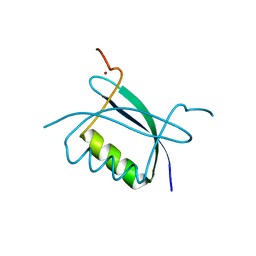 | | Crystal Structure of HPV E7 CR3 domain | | Descriptor: | E7 protein, ZINC ION | | Authors: | Liu, X, Clements, A, Zhao, K, Marmorstein, R. | | Deposit date: | 2005-10-11 | | Release date: | 2005-10-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the human Papillomavirus E7 oncoprotein and its mechanism for inactivation of the retinoblastoma tumor suppressor.
J.Biol.Chem., 281, 2006
|
|
5HQT
 
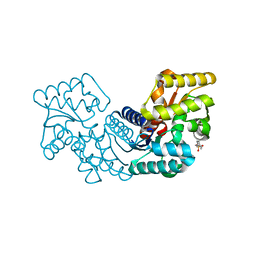 | | Crystal structure of an aspartate/glutamate racemase from Escherichia coli O157 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, aspartate/glutamate racemase | | Authors: | Liu, X, Gao, F, Ma, Y, Liu, S, Cui, Y, Yuan, Z, Kang, X. | | Deposit date: | 2016-01-22 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Crystal structure and molecular mechanism of an aspartate/glutamate racemase from Escherichia coli O157
Febs Lett., 590, 2016
|
|
2YXP
 
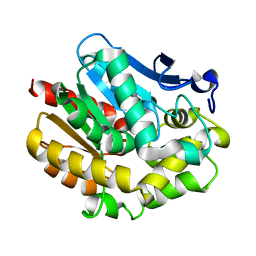 | | The Effect of Deuteration on Protein Structure A High Resolution Comparison of Hydrogenous and Perdeuterated Haloalkane Dehalogenase | | Descriptor: | Haloalkane dehalogenase | | Authors: | Liu, X, Hanson, L, Langan, P, Viola, R.E. | | Deposit date: | 2007-04-27 | | Release date: | 2007-09-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | The effect of deuteration on protein structure: a high-resolution comparison of hydrogenous and perdeuterated haloalkane dehalogenase.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
3BIY
 
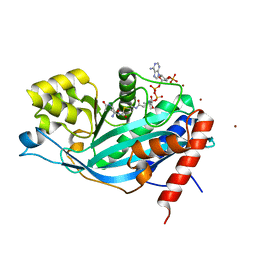 | | Crystal structure of p300 histone acetyltransferase domain in complex with a bisubstrate inhibitor, Lys-CoA | | Descriptor: | BROMIDE ION, Histone acetyltransferase p300, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]methyl (3R,20R)-20-carbamoyl-3-hydroxy-2,2-dimethyl-4,8,14,22-tetraoxo-12-thia-5,9,15,21-tetraazatricos-1-yl dihydrogen diphosphate | | Authors: | Liu, X, Wang, L, Zhao, K, Thompson, P.R, Hwang, Y, Marmorstein, R, Cole, P.A. | | Deposit date: | 2007-12-02 | | Release date: | 2008-02-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structural basis of protein acetylation by the p300/CBP transcriptional coactivator
Nature, 451, 2008
|
|
3C20
 
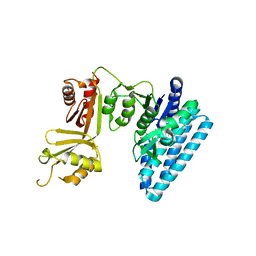 | |
3C1M
 
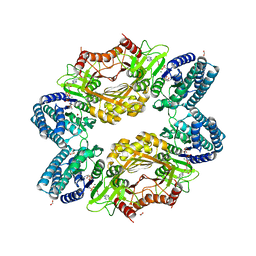 | |
3C1N
 
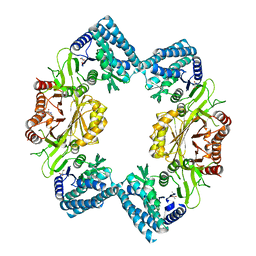 | |
1PQ1
 
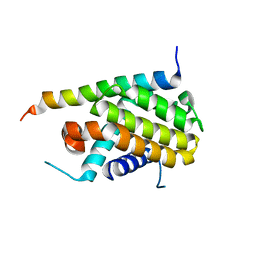 | | Crystal structure of Bcl-xl/Bim | | Descriptor: | Apoptosis regulator Bcl-X, BCL2-like protein 11 | | Authors: | Liu, X, Dai, S, Zhu, Y, Marrack, P, Kappler, J.W. | | Deposit date: | 2003-06-17 | | Release date: | 2003-09-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The structure of a Bcl-xl/Bim fragment complex: Implications for Bim function
Immunity, 19, 2003
|
|
1PQ0
 
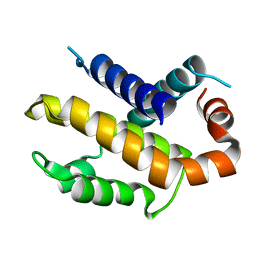 | | Crystal structure of mouse Bcl-xl | | Descriptor: | Apoptosis regulator Bcl-X | | Authors: | Liu, X, Dai, S, Zhu, Y, Marrack, P, Kappler, J.W. | | Deposit date: | 2003-06-17 | | Release date: | 2003-09-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of a Bcl-xl/Bim fragment complex: Implications for Bim function
Immunity, 19, 2003
|
|
8DQL
 
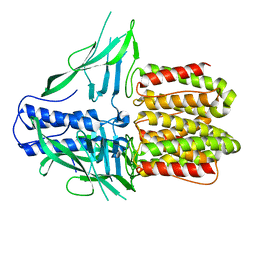 | | CryoEM structure of IglD | | Descriptor: | Secretion system protein | | Authors: | Liu, X, Clemens, D, Lee, B, Yang, X, Zhou, H, Horwitz, M. | | Deposit date: | 2022-07-19 | | Release date: | 2022-08-17 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Atomic Structure of IglD Demonstrates Its Role as a Component of the Baseplate Complex of the Francisella Type VI Secretion System.
Mbio, 13, 2022
|
|
6AAX
 
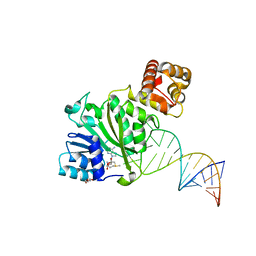 | | Crystal structure of TFB1M and h45 with SAM in homo sapiens | | Descriptor: | DI(HYDROXYETHYL)ETHER, Dimethyladenosine transferase 1, mitochondrial, ... | | Authors: | Liu, X, Shen, S, Wu, P, Li, F, Gong, Q, Wu, J, Zhang, H, Shi, Y. | | Deposit date: | 2018-07-19 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.994 Å) | | Cite: | Structural insights into dimethylation of 12S rRNA by TFB1M: indispensable role in translation of mitochondrial genes and mitochondrial function.
Nucleic Acids Res., 47, 2019
|
|
6AAU
 
 | | Solution Structure for m62A helix 45 in 3' end of 12S rRNA | | Descriptor: | RNA (24-mer) | | Authors: | Liu, X, Wu, P. | | Deposit date: | 2018-07-19 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insights into dimethylation of 12S rRNA by TFB1M: indispensable role in translation of mitochondrial genes and mitochondrial function.
Nucleic Acids Res., 47, 2019
|
|
2R7G
 
 | |
