7ZXK
 
 | | Human IL-27 in complex with neutralizing SRF388 FAb fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-27 subunit alpha, Interleukin-27 subunit beta, ... | | Authors: | Bloch, Y, Skladanowska, K, Strand, J, Welin, M, Logan, D, Hill, J, Savvides, S.N. | | Deposit date: | 2022-05-21 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of activation and antagonism of receptor signaling mediated by interleukin-27.
Cell Rep, 41, 2022
|
|
1KQL
 
 | | Crystal structure of the C-terminal region of striated muscle alpha-tropomyosin at 2.7 angstrom resolution | | Descriptor: | Fusion Protein of and striated muscle alpha-tropomyosin and the GCN4 leucine zipper | | Authors: | Li, Y, Mui, S, Brown, J.H, Strand, J, Reshetnikova, L, Tobacman, L.S, Cohen, C. | | Deposit date: | 2002-01-07 | | Release date: | 2002-05-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of the C-terminal fragment of striated-muscle alpha-tropomyosin reveals a key troponin T recognition site.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
7PFL
 
 | | The SARS-CoV2 major protease (Mpro) apo structure to 1.8 A resolution | | Descriptor: | Replicase polyprotein 1ab, SODIUM ION | | Authors: | Moche, M, Moodie, L, Strandback, E, Nyman, T, Akaberi, D, Lennerstrand, J. | | Deposit date: | 2021-08-11 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The SARS-CoV2 major protease (Mpro) in complex with a non-covalent inhibitory ligand at 2 A resolution
To Be Published
|
|
7PFM
 
 | | A SARS-CoV2 major protease non-covalent ligand structure determined to 2.0 A resolution | | Descriptor: | N-[(1R)-2-(tert-butylamino)-2-oxidanylidene-1-pyridin-3-yl-ethyl]-N-(4-tert-butylphenyl)-1H-imidazole-5-carboxamide, Replicase polyprotein 1ab | | Authors: | Moche, M, Moodie, L, Strandback, E, Nyman, T, Sandstrom, A, Akaberi, D, Lennerstrand, J. | | Deposit date: | 2021-08-11 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The SARS-CoV2 major protease (Mpro) in complex with a non-covalent inhibitory ligand at 2 A resolution
To Be Published
|
|
2K3S
 
 | | HADDOCK-derived structure of the CH-domain of the smoothelin-like 1 complexed with the C-domain of apocalmodulin | | Descriptor: | Calmodulin, Smoothelin-like protein 1 | | Authors: | Ishida, H, Borman, M.A, Ostrander, J, Vogel, H.J, MacDonald, J.A. | | Deposit date: | 2008-05-15 | | Release date: | 2008-05-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the calponin homology (CH) domain from the smoothelin-like 1 protein: a unique apocalmodulin-binding mode and the possible role of the C-terminal type-2 CH-domain in smooth muscle relaxation.
J.Biol.Chem., 283, 2008
|
|
7ZG0
 
 | | Murine IL-27 in complex with IL-27Ra and a non-competing Nb | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-27 receptor subunit alpha, ... | | Authors: | Skladanowska, K, Bloch, Y, Savvides, S.N. | | Deposit date: | 2022-04-01 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Structural basis of activation and antagonism of receptor signaling mediated by interleukin-27.
Cell Rep, 41, 2022
|
|
3RME
 
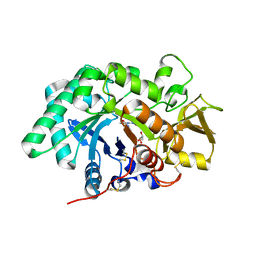 | | AMCase in complex with Compound 5 | | Descriptor: | Acidic mammalian chitinase, GLYCEROL, N-ethyl-2-(4-methylpiperazin-1-yl)pyridine-3-carboxamide | | Authors: | Olland, A. | | Deposit date: | 2011-04-20 | | Release date: | 2011-08-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification and Characterization of Acidic Mammalian Chitinase Inhibitors
J.Med.Chem., 53, 2010
|
|
3RM8
 
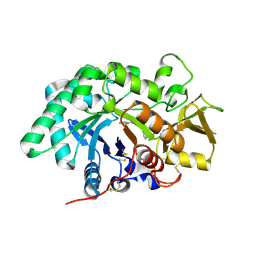 | | AMCase in complex with Compound 2 | | Descriptor: | 2-methyl-3-{[4-(pyridin-2-yl)piperazin-1-yl]methyl}-1H-indole, Acidic mammalian chitinase | | Authors: | Olland, A. | | Deposit date: | 2011-04-20 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification and Characterization of Acidic Mammalian Chitinase Inhibitors
J.Med.Chem., 53, 2010
|
|
3RM9
 
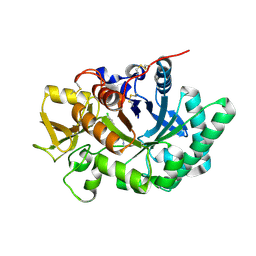 | | AMCase in complex with Compound 3 | | Descriptor: | 4-(4-chlorophenyl)piperazine-1-carboximidamide, Acidic mammalian chitinase | | Authors: | Olland, A. | | Deposit date: | 2011-04-20 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification and Characterization of Acidic Mammalian Chitinase Inhibitors
J.Med.Chem., 53, 2010
|
|
3RM4
 
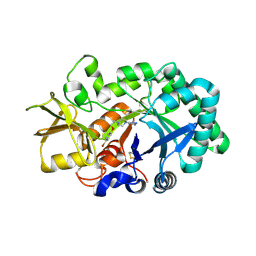 | | AMCase in complex with Compound 1 | | Descriptor: | 5-{4-[2-(4-bromophenoxy)ethyl]piperazin-1-yl}-4H-1,2,4-triazol-3-amine, Acidic mammalian chitinase | | Authors: | Olland, A. | | Deposit date: | 2011-04-20 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification and Characterization of Acidic Mammalian Chitinase Inhibitors
J.Med.Chem., 53, 2010
|
|
3FXY
 
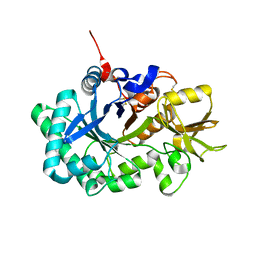 | | Acidic Mammalian Chinase, Catalytic Domain | | Descriptor: | Acidic mammalian chitinase | | Authors: | Olland, A.M. | | Deposit date: | 2009-01-21 | | Release date: | 2009-03-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Triad of polar residues implicated in pH specificity of acidic mammalian chitinase.
Protein Sci., 18, 2009
|
|
3FY1
 
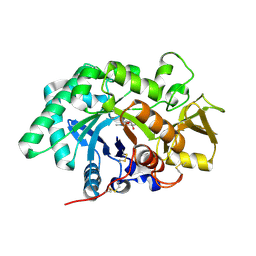 | | The Acidic Mammalian Chitinase catalytic domain in complex with methylallosamidin | | Descriptor: | 2-acetamido-2-deoxy-6-O-methyl-alpha-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-allopyranose, ALLOSAMIZOLINE, Acidic mammalian chitinase | | Authors: | Olland, A.M. | | Deposit date: | 2009-01-21 | | Release date: | 2009-03-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Triad of polar residues implicated in pH specificity of acidic mammalian chitinase.
Protein Sci., 18, 2009
|
|
7B2U
 
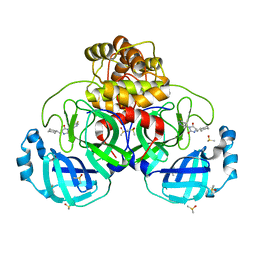 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 1 | | Descriptor: | (5S)-5-(cyclohexylmethyl)-3-(5-fluoropyridin-3-yl)imidazolidine-2,4-dione, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Talibov, V.O. | | Deposit date: | 2020-11-27 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
7B77
 
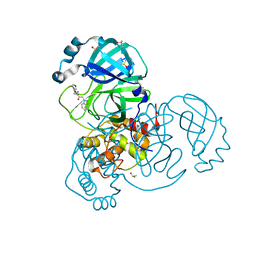 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 8 | | Descriptor: | 2-(benzotriazol-1-yl)-~{N}-ethyl-~{N}-(furan-3-ylmethyl)ethanamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Talibov, V.O. | | Deposit date: | 2020-12-09 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
7B5Z
 
 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 6 | | Descriptor: | 2-(1H-benzo[d][1,2,3]triazol-1-yl)-1-(4-methylenepiperidin-1-yl)ethan-1-one, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Talibov, V.O. | | Deposit date: | 2020-12-07 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
7AU4
 
 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 3 | | Descriptor: | (3~{S})-6-chloranyl-3'-(1,2-oxazol-3-ylmethyl)spiro[1,2-dihydroindene-3,5'-imidazolidine]-2',4'-dione, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Talibov, V.O. | | Deposit date: | 2020-11-02 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
7B2J
 
 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 5 | | Descriptor: | 2-(1H-1,2,3-benzotriazol-1-yl)-1-(4-methylpiperidin-1-yl)ethan-1-one, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Talibov, V.O. | | Deposit date: | 2020-11-27 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
7QBB
 
 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 18 | | Descriptor: | 3C-like proteinase nsp5, 7-isoquinolin-4-yl-2-phenyl-5,7-diazaspiro[3.4]octane-6,8-dione, DIMETHYL SULFOXIDE | | Authors: | Talibov, V.O. | | Deposit date: | 2021-11-18 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
7NEO
 
 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 15 | | Descriptor: | 2-cyclobutyl-7-(5-fluoropyridin-3-yl)-5,7-diazaspiro[3.4]octane-6,8-dione, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Talibov, V.O. | | Deposit date: | 2021-02-04 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
7NBT
 
 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 21 | | Descriptor: | 2-(benzotriazol-1-yl)-1-[(4~{S})-4-methyl-6,7-dihydro-4~{H}-thieno[3,2-c]pyridin-5-yl]ethanone, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Talibov, V.O. | | Deposit date: | 2021-01-27 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
7O46
 
 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 17 | | Descriptor: | 2-cyclobutyl-7-isoquinolin-4-yl-5,7-diazaspiro[3.4]octane-6,8-dione, 3C-like proteinase nsp5 | | Authors: | Talibov, V.O. | | Deposit date: | 2021-04-05 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
7BIJ
 
 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 13 | | Descriptor: | (3~{S})-3'-(5-fluoranylpyridin-3-yl)spiro[1,2-dihydroindene-3,5'-imidazolidine]-2',4'-dione, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Talibov, V.O. | | Deposit date: | 2021-01-12 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
8C1U
 
 | | SFX structure of D.m(6-4)photolyase | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8C69
 
 | | Light SFX structure of D.m(6-4)photolyase at 100 microsecond time delay | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8C6A
 
 | | Light SFX structure of D.m(6-4)photolyase at 1ps time delay | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
