7MEZ
 
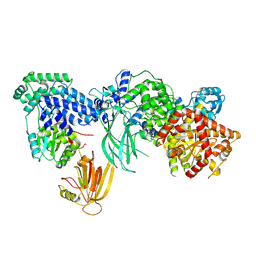 | | Structure of the phosphoinositide 3-kinase p110 gamma (PIK3CG) p101 (PIK3R5) complex | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, Phosphoinositide 3-kinase regulatory subunit 5 | | Authors: | Burke, J.E, Dalwadi, U, Rathinaswamy, M.K, Yip, C.K. | | Deposit date: | 2021-04-08 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure of the phosphoinositide 3-kinase (PI3K) p110 gamma-p101 complex reveals molecular mechanism of GPCR activation.
Sci Adv, 7, 2021
|
|
7JX0
 
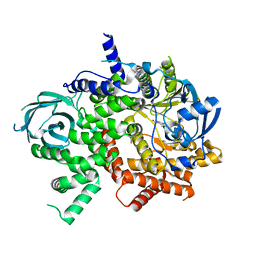 | | NVS-PI3-4 bound to the PI3Kg catalytic subunit p110 gamma | | Descriptor: | N~3~-{[5-(4-acetylphenyl)-4-methyl-1,3-thiazol-2-yl]carbamoyl}-N-tert-butyl-beta-alaninamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Burke, J.E, Rathinaswamy, M.K, Harris, N.J. | | Deposit date: | 2020-08-26 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Disease related mutations in PI3K gamma disrupt regulatory C-terminal dynamics and reveal a path to selective inhibitors.
Elife, 10, 2021
|
|
7JWE
 
 | |
7JWZ
 
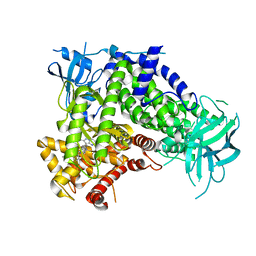 | | IPI-549 bound to the PI3Kg catalytic subunit p110 gamma | | Descriptor: | 2-amino-N-[(1S)-1-{8-[(1-methyl-1H-pyrazol-4-yl)ethynyl]-1-oxo-2-phenyl-1,2-dihydroisoquinolin-3-yl}ethyl]pyrazolo[1,5-a]pyrimidine-3-carboxamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Burke, J.E, Rathinaswamy, M.K, Harris, N.J. | | Deposit date: | 2020-08-26 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Disease related mutations in PI3K gamma disrupt regulatory C-terminal dynamics and reveal a path to selective inhibitors.
Elife, 10, 2021
|
|
8DP0
 
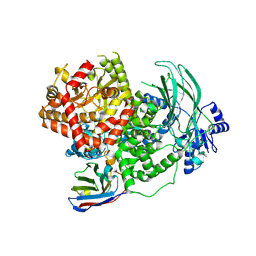 | | Structure of p110 gamma bound to the Ras inhibitory nanobody NB7 | | Descriptor: | Nanobody NB7, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Burke, J.E, Nam, S.E, Rathinaswamy, M.K, Yip, C.K. | | Deposit date: | 2022-07-14 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Allosteric activation or inhibition of PI3K gamma mediated through conformational changes in the p110 gamma helical domain.
Elife, 12, 2023
|
|
8AJ8
 
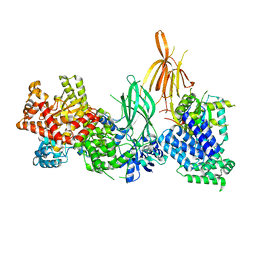 | | Structure of p110 gamma bound to the p84 regulatory subunit | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, Phosphoinositide 3-kinase regulatory subunit 6 | | Authors: | Burke, J.E, Williams, R.L, Zhang, X. | | Deposit date: | 2022-07-27 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (8.5 Å) | | Cite: | Molecular basis for differential activation of p101 and p84 complexes of PI3K gamma by Ras and GPCRs.
Cell Rep, 42, 2023
|
|
8U0H
 
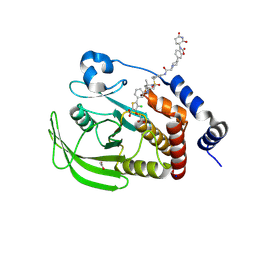 | | Crystal structure of PTPN2 with a PROTAC | | Descriptor: | (5P)-3-(carboxymethoxy)-4-chloro-5-(3-{[(4S)-1-({3-[2-(4-{3-[(3R)-2,6-dioxopiperidin-3-yl]-2-oxo-2,3-dihydro-1,3-benzoxazol-6-yl}piperidin-1-yl)acetamido]phenyl}methanesulfonyl)-2,2-dimethylpiperidin-4-yl]amino}phenyl)thiophene-2-carboxylic acid, ACETATE ION, PTPN2, ... | | Authors: | Jain, R, Longenecker, K, Qiu, W. | | Deposit date: | 2023-08-29 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Mechanistic insights into a heterobifunctional degrader-induced PTPN2/N1 complex.
Commun Chem, 7, 2024
|
|
8UH6
 
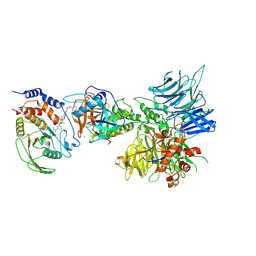 | | Degrader-induced complex between PTPN2 and CRBN-DDB1 | | Descriptor: | (5P)-3-(carboxymethoxy)-4-chloro-5-(3-{[(4S)-1-({3-[(4-{1-[(3R)-2,6-dioxopiperidin-3-yl]-3-methyl-2-oxo-2,3-dihydro-1H-benzimidazol-5-yl}piperidine-1-carbonyl)amino]phenyl}methanesulfonyl)-2,2-dimethylpiperidin-4-yl]amino}phenyl)thiophene-2-carboxylic acid, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Catalano, C, Bratkowski, M, Scapin, G, Hao, Q. | | Deposit date: | 2023-10-06 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanistic insights into a heterobifunctional degrader-induced PTPN2/N1 complex.
Commun Chem, 7, 2024
|
|
8VU5
 
 | |
5VBE
 
 | | Crystal Structure of Small Molecule Disulfide 2C07 Bound to H-Ras M72C GDP | | Descriptor: | 1-(4-methoxyphenyl)-N-(3-sulfanylpropyl)-5-(trifluoromethyl)-1H-pyrazole-4-carboxamide, GTPase HRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gentile, D.R, Jenkins, M.L, Moss, S.M, Burke, J.E, Shokat, K.M. | | Deposit date: | 2017-03-29 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Ras Binder Induces a Modified Switch-II Pocket in GTP and GDP States.
Cell Chem Biol, 24, 2017
|
|
5VBZ
 
 | | Crystal Structure of Small Molecule Disulfide 2C07 Bound to H-Ras M72C GppNHp | | Descriptor: | 1-(4-methoxyphenyl)-N-(3-sulfanylpropyl)-5-(trifluoromethyl)-1H-pyrazole-4-carboxamide, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Gentile, D.R, Jenkins, M.L, Moss, S.M, Burke, J.E, Shokat, K.M. | | Deposit date: | 2017-03-30 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ras Binder Induces a Modified Switch-II Pocket in GTP and GDP States.
Cell Chem Biol, 24, 2017
|
|
5VBM
 
 | | Crystal Structure of Small Molecule Disulfide 2C07 Bound to K-Ras Cys Light M72C GDP | | Descriptor: | 1-(4-methoxyphenyl)-N-(3-sulfanylpropyl)-5-(trifluoromethyl)-1H-pyrazole-4-carboxamide, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gentile, D.R, Jenkins, M.L, Moss, S.M, Burke, J.E, Shokat, K.M. | | Deposit date: | 2017-03-29 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Ras Binder Induces a Modified Switch-II Pocket in GTP and GDP States.
Cell Chem Biol, 24, 2017
|
|
