4IJO
 
 | | Unraveling hidden allosteric regulatory sites in structurally homologues metalloproteases | | Descriptor: | CALCIUM ION, Macrophage metalloelastase, ZINC ION | | Authors: | Udi, Y, Fragai, M, Grossman, M, Mitternacht, S, Arad-Yellin, R, Calderone, V, Melikian, M, Toccafondi, M, Berezovsky, I.N, Luchinat, C, Sagi, I. | | Deposit date: | 2012-12-22 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unraveling hidden regulatory sites in structurally homologous metalloproteases
J.Mol.Biol., 425, 2013
|
|
6TI5
 
 | | A New Structural Model of Abeta(1-40) Fibrils | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Bertini, I, Gonnelli, L, Luchinat, C, Mao, J, Nesi, A. | | Deposit date: | 2019-11-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | Mixing A beta (1-40) and A beta (1-42) peptides generates unique amyloid fibrils.
Chem.Commun.(Camb.), 56, 2020
|
|
2B1U
 
 | | Solution structure of Calmodulin-like Skin Protein C terminal domain | | Descriptor: | Calmodulin-like protein 5 | | Authors: | Babini, E, Bertini, I, Capozzi, F, Chirivino, E, Luchinat, C, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-09-16 | | Release date: | 2006-05-30 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A Structural and Dynamic Characterization of the EF-Hand Protein CLSP.
Structure, 14, 2006
|
|
6EOK
 
 | | Crystal structure of E. coli L-asparaginase II | | Descriptor: | L-asparaginase 2, ZINC ION | | Authors: | Cerofolini, L, Giuntini, S, Carlon, A, Ravera, E, Calderone, V, Fragai, M, Parigi, G, Luchinat, C. | | Deposit date: | 2017-10-09 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Characterization of PEGylated Asparaginase: New Opportunities from NMR Analysis of Large PEGylated Therapeutics.
Chemistry, 25, 2019
|
|
6YEL
 
 | | Stromal interaction molecule 1 coiled-coil 1 fragment | | Descriptor: | Stromal interaction molecule 1 | | Authors: | Rathner, P, Cerofolini, L, Ravera, E, Bechmann, M, Grabmayr, H, Fahrner, M, Fragai, M, Romanin, C, Luchinat, C, Mueller, N. | | Deposit date: | 2020-03-25 | | Release date: | 2020-09-02 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Interhelical interactions within the STIM1 CC1 domain modulate CRAC channel activation.
Nat.Chem.Biol., 17, 2021
|
|
1RJU
 
 | | Crystal structure of a truncated form of yeast copper thionein | | Descriptor: | COPPER (I) ION, Metallothionein | | Authors: | Calderone, V, Dolderer, B, Hartmann, H.J, Echner, H, Luchinat, C, Del Bianco, C, Mangani, S, Weser, U. | | Deposit date: | 2003-11-20 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | The Crystal Structure of Yeast Copper Thionein: the solution of a long lasting Enigma.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1CLF
 
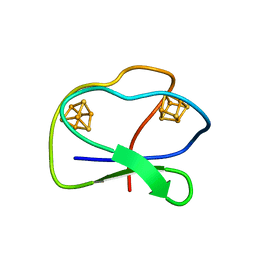 | | CLOSTRIDIUM PASTEURIANUM FERREDOXIN | | Descriptor: | FERREDOXIN, IRON/SULFUR CLUSTER | | Authors: | Bertini, I, Donaire, A, Feinberg, B.A, Luchinat, C, Piccioli, M, Yuan, H. | | Deposit date: | 1995-06-21 | | Release date: | 1996-01-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the oxidized 2[4Fe-4S] ferredoxin from Clostridium pasteurianum.
Eur.J.Biochem., 232, 1995
|
|
1Y93
 
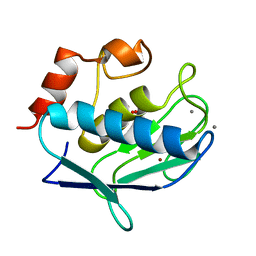 | | Crystal structure of the catalytic domain of human MMP12 complexed with acetohydroxamic acid at atomic resolution | | Descriptor: | ACETOHYDROXAMIC ACID, CALCIUM ION, Macrophage metalloelastase, ... | | Authors: | Bertini, I, Calderone, V, Cosenza, M, Fragai, M, Lee, Y.-M, Luchinat, C, Mangani, S, Terni, B, Turano, P. | | Deposit date: | 2004-12-14 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Conformational variability of matrix metalloproteinases: Beyond a single 3D structure
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
3RTT
 
 | | Human MMP-12 catalytic domain in complex with*(R)-N*-Hydroxy-1-(phenethylsulfonyl)pyrrolidine-2-carboxamide | | Descriptor: | CALCIUM ION, Macrophage metalloelastase, N-hydroxy-1-[(2-phenylethyl)sulfonyl]-D-prolinamide, ... | | Authors: | Bertini, I, Calderone, V, Fragai, M, Luchinat, C, Mori, M, Nativi, C. | | Deposit date: | 2011-05-04 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Contribution of ligand free energy of solvation to design new potent MMPs inhibitors.
J.Med.Chem., 2012
|
|
3RTS
 
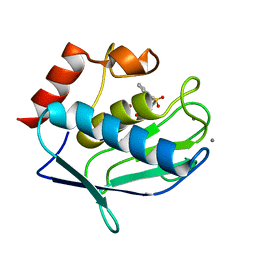 | | Human MMP-12 catalytic domain in complex with*N*-Hydroxy-2-(2-phenylethylsulfonamido)acetamide | | Descriptor: | CALCIUM ION, Macrophage metalloelastase, N-hydroxy-N~2~-[(2-phenylethyl)sulfonyl]glycinamide, ... | | Authors: | Bertini, I, Calderone, V, Fragai, M, Luchinat, C, Mori, M, Nativi, C. | | Deposit date: | 2011-05-04 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Contribution of ligand free energy of solvation to design new potent MMPs inhibitors.
J.Med.Chem., 2012
|
|
6TI6
 
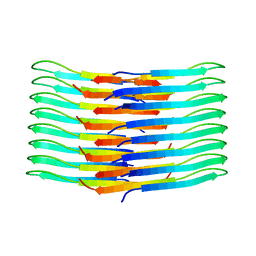 | | Mixing Abeta(1-40) and Abeta(1-42) peptides generates unique amyloid fibrils | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Cerofolini, L, Ravera, E, Bologna, S, Wiglenda, T, Boddrich, A, Purfurst, B, Benilova, A, Korsak, M, Gallo, G, Rizzo, D, Gonnelli, L, Fragai, M, De Strooper, B, Wanker, E.E, Luchinat, C. | | Deposit date: | 2019-11-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | Mixing A beta (1-40) and A beta (1-42) peptides generates unique amyloid fibrils.
Chem.Commun.(Camb.), 56, 2020
|
|
7A70
 
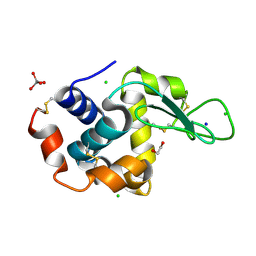 | | HEW lysozyme in complex with Ti(OH)4 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme, ... | | Authors: | Calderone, V, Gigli, L, Ravera, E, Luchinat, C. | | Deposit date: | 2020-08-27 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | On the Mechanism of Bioinspired Formation of Inorganic Oxides: Structural Evidence of the Electrostatic Nature of the Interaction between a Mononuclear Inorganic Precursor and Lysozyme.
Biomolecules, 11, 2020
|
|
6H6S
 
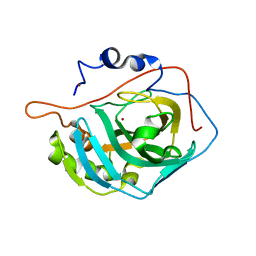 | | Sad phasing on nickel-substituted human carbonic anhydrase II | | Descriptor: | Carbonic anhydrase 2, NICKEL (II) ION | | Authors: | Calderone, V, Fragai, M, Silva, J.P, Luchinat, C, Ravera, E, Geraldes, C.F.G.C, Macedo, A.L, Cerofolini, L, Giuntini, S. | | Deposit date: | 2018-07-30 | | Release date: | 2019-01-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Non-crystallographic symmetry in proteins: Jahn-Teller-like and Butterfly-like effects?
J. Biol. Inorg. Chem., 24, 2019
|
|
3BA0
 
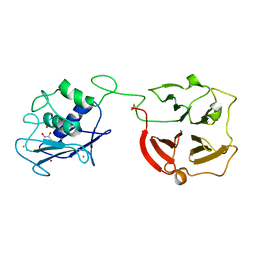 | | Crystal structure of full-length human MMP-12 | | Descriptor: | ACETOHYDROXAMIC ACID, CALCIUM ION, Macrophage metalloelastase, ... | | Authors: | Bertini, I, Calderone, V, Fragai, M, Jaiswal, R, Luchinat, C, Melikian, M, Myonas, E, Svergun, D.I. | | Deposit date: | 2007-11-07 | | Release date: | 2008-07-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Evidence of reciprocal reorientation of the catalytic and hemopexin-like domains of full-length MMP-12.
J.Am.Chem.Soc., 130, 2008
|
|
5LAB
 
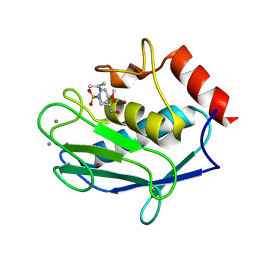 | |
4GUY
 
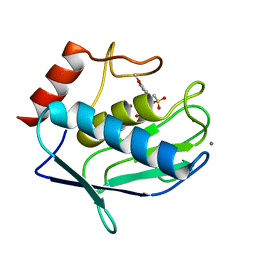 | | Human MMP12 catalytic domain in complex with*N*-Hydroxy-2-(2-(4-methoxyphenyl)ethylsulfonamido)acetamide | | Descriptor: | CALCIUM ION, Macrophage metalloelastase, N-hydroxy-N~2~-{[2-(4-methoxyphenyl)ethyl]sulfonyl}glycinamide, ... | | Authors: | Calderone, V, Fragai, M, Luchinat, C, Massaro, A, Mordini, A, Mori, M. | | Deposit date: | 2012-08-30 | | Release date: | 2012-09-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Contribution of free energy of solvation to ligand affinity in new potent MMPs inhibitors.
To be Published
|
|
1NOE
 
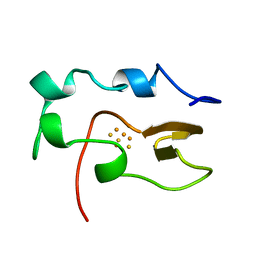 | | NMR STUDY OF REDUCED HIGH POTENTIAL IRON SULFUR PROTEIN | | Descriptor: | HIGH POTENTIAL IRON SULFUR PROTEIN, IRON/SULFUR CLUSTER | | Authors: | Bentrop, D, Bertini, I, Capozzi, F, Dikiy, A, Eltis, L, Luchinat, C. | | Deposit date: | 1996-01-07 | | Release date: | 1996-06-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the reduced C77S mutant of the Chromatium vinosum high-potential iron-sulfur protein through nuclear magnetic resonance: comparison with the solution structure of the wild-type protein.
Biochemistry, 35, 1996
|
|
8AWW
 
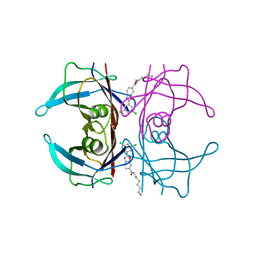 | | Transthyretin conjugated with a tafamidis derivative | | Descriptor: | Transthyretin, ~{N}-(6-azanylhexyl)-2-[3,5-bis(chloranyl)phenyl]-1,3-benzoxazole-6-carboxamide | | Authors: | Cerofolini, L, Vasa, K, Bianconi, E, Salobehaj, M, Cappelli, G, Licciardi, G, Perez-Rafols, A, Padilla Cortes, L.D, Antonacci, S, Rizzo, D, Ravera, E, Calderone, V, Parigi, G, Luchinat, C, Macchiarulo, A, Menichetti, S, Fragai, M. | | Deposit date: | 2022-08-30 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Combining Solid-State NMR with Structural and Biophysical Techniques to Design Challenging Protein-Drug Conjugates.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
1RMZ
 
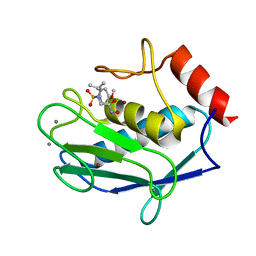 | | Crystal structure of the catalytic domain of human MMP12 complexed with the inhibitor NNGH at 1.3 A resolution | | Descriptor: | CALCIUM ION, Macrophage metalloelastase, N-ISOBUTYL-N-[4-METHOXYPHENYLSULFONYL]GLYCYL HYDROXAMIC ACID, ... | | Authors: | Bertini, I, Calderone, V, Fragai, M, Luchinat, C, Mangani, S, Terni, B. | | Deposit date: | 2003-11-28 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Conformational variability of matrix metalloproteinases: beyond a single 3D structure.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
5HYD
 
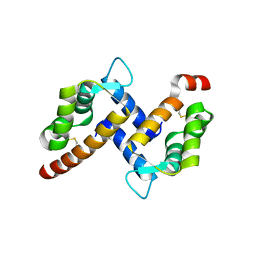 | |
8OGD
 
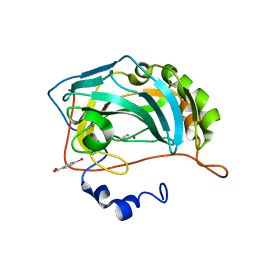 | | Structure of zinc(II) double mutant human carbonic anhydrase II bound to thiocyanate | | Descriptor: | 4-(HYDROXYMERCURY)BENZOIC ACID, Carbonic anhydrase 2, THIOCYANATE ION, ... | | Authors: | Silva, J.M, Cerofolini, L, Carvalho, A.L, Ravera, E, Fragai, M, Parigi, G, Macedo, A.L, Geraldes, C.F.G.C, Luchinat, C. | | Deposit date: | 2023-03-20 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Elucidating the concentration-dependent effects of thiocyanate binding to carbonic anhydrase.
J.Inorg.Biochem., 244, 2023
|
|
8OGE
 
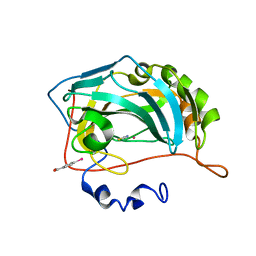 | | Structure of cobalt(II) substituted double mutant human carbonic anhydrase II bound to thiocyanate | | Descriptor: | 4-(HYDROXYMERCURY)BENZOIC ACID, COBALT (II) ION, Carbonic anhydrase 2, ... | | Authors: | Silva, J.M, Cerofolini, L, Carvalho, A.L, Ravera, E, Fragai, M, Parigi, G, Macedo, A.L, Geraldes, C.F.G.C, Luchinat, C. | | Deposit date: | 2023-03-20 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Elucidating the concentration-dependent effects of thiocyanate binding to carbonic anhydrase.
J.Inorg.Biochem., 244, 2023
|
|
1NEH
 
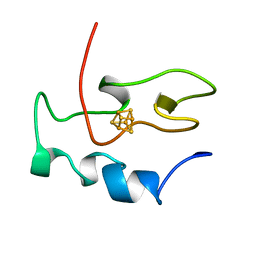 | | HIGH POTENTIAL IRON-SULFUR PROTEIN | | Descriptor: | HIGH POTENTIAL IRON SULFUR PROTEIN, IRON/SULFUR CLUSTER | | Authors: | Bertini, I, Dikiy, A, Kastrau, D.H.W, Luchinat, C, Sompornpisut, P. | | Deposit date: | 1995-12-14 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the oxidized high potential iron-sulfur protein from Chromatium vinosum through NMR. Comparative analysis with the solution structure of the reduced species.
Biochemistry, 34, 1995
|
|
1JXD
 
 | | SOLUTION STRUCTURE OF REDUCED CU(I) PLASTOCYANIN FROM SYNECHOCYSTIS PCC6803 | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Bertini, I, Bryant, D.A, Ciurli, S, Dikiy, A, Fernandez, C.O, Luchinat, C, Safarov, N, Vila, A.J, Zhao, J. | | Deposit date: | 2001-09-07 | | Release date: | 2001-09-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Backbone dynamics of plastocyanin in both oxidation states. Solution structure of the reduced form and comparison with the oxidized state.
J.Biol.Chem., 276, 2001
|
|
1HRQ
 
 | | THE THREE-DIMENSIONAL SOLUTION STRUCTURE OF THE REDUCED HIGH-POTENTIAL IRON-SULFUR PROTEIN FROM CHROMATIUM VINOSUM THROUGH NMR | | Descriptor: | HIGH POTENTIAL IRON SULFUR PROTEIN, IRON/SULFUR CLUSTER | | Authors: | Banci, L, Bertini, I, Dikiy, A, Kastrau, D.H.W, Luchinat, C, Sompornpisut, P. | | Deposit date: | 1995-01-17 | | Release date: | 1995-06-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional solution structure of the reduced high-potential iron-sulfur protein from Chromatium vinosum through NMR.
Biochemistry, 34, 1995
|
|
