5F4Z
 
 | | The crystal structure of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus | | Descriptor: | (1~{R},2~{R})-2,3-dihydro-1~{H}-indene-1,2-diol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Tan, K, Li, H, Jedrzejczak, R, BABNIGG, G, BINGMAN, C.A, YENNAMALLI, R, LOHMAN, J, Chang, C.Y, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-12-03 | | Release date: | 2016-02-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The crystal structure of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus
To Be Published
|
|
4QA9
 
 | | Ensemble refinement of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus. | | Descriptor: | 1,2-ETHANEDIOL, Epoxide hydrolase, SULFATE ION | | Authors: | Wang, F, Tan, K, Bigelow, L, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Joachimiak, A, Phillips Jr, G.N, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-05-02 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Ensemble refinement of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus.
To be Published
|
|
4Z5P
 
 | | Crystal structure of the LnmA cytochrome P450 hydroxylase from the leinamycin biosynthetic pathway of Streptomyces atroolivaceus S-140 at 1.9 A resolution | | Descriptor: | Cytochrome P450 hydroxylase, PROTOPORPHYRIN IX CONTAINING FE, TRIETHYLENE GLYCOL | | Authors: | Ma, M, Lohman, J, Rudolf, J, Miller, M.D, Cao, H, Osipiuk, J, Babnigg, G, Phillips Jr, G.N, Joachimiak, A, Shen, B, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-04-02 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the LnmA cytochrome P450 hydroxylase from the leinamycin biosynthetic pathway of Streptomyces atroolivaceus S-140
To be Published
|
|
4Z5Q
 
 | | Crystal structure of the LnmZ cytochrome P450 hydroxylase from the leinamycin biosynthetic pathway of Streptomyces atroolivaceus S-140 at 1.8 A resolution | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, Cytochrome P450 hydroxylase, ... | | Authors: | Ma, M, Lohman, J, Rudolf, J, Miller, M.D, Cao, H, Osipiuk, J, Joachimiak, A, Phillips Jr, G.N, Shen, B, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-04-02 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal structure of the LnmZ cytochrome P450 hydroxylase from the leinamycin biosynthetic pathway of Streptomyces atroolivaceus S-140
To be Published
|
|
5BMO
 
 | | LnmX protein, a putative GlcNAc-PI de-N-acetylase from Streptomyces atroolivaceus | | Descriptor: | ACETATE ION, POTASSIUM ION, Putative uncharacterized protein LnmX | | Authors: | Osipiuk, J, Hatzos-Skintges, C, Cuff, M, Endres, M, Babnigg, G, Lohman, J, Ma, M, Rudolf, J, Chang, C.-Y, Shen, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-05-22 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | LnmX protein, a putative GlcNAc-PI de-N-acetylase from Streptomyces atroolivaceus.
to be published
|
|
4ZNM
 
 | | Crystal structure of SgcC5 protein from Streptomyces globisporus (apo form) | | Descriptor: | C-domain type II peptide synthetase, CHLORIDE ION, SODIUM ION | | Authors: | Michalska, K, Bigelow, L, Jedrzejczak, R, Babnigg, G, Lohman, J, Ma, M, Rudolf, J, Chang, C.-Y, Shen, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-05-04 | | Release date: | 2015-05-27 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Crystal structure of SgcC5 protein from Streptomyces globisporus (apo form)
To Be Published
|
|
4ZXW
 
 | | Crystal structure of SgcC5 protein from Streptomyces globisporus (complex with (R)-(-)-1-(2-naphthyl)-1,2-ethanediol and sucrose) | | Descriptor: | (1R)-1-(naphthalen-2-yl)ethane-1,2-diol, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, C-domain type II peptide synthetase, ... | | Authors: | Michalska, K, Bigelow, L, Jedrzejczak, R, Babnigg, G, Lohman, J, Ma, M, Rudolf, J, Chang, C.-Y, Shen, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-05-20 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | Crystal structure of SgcC5 protein from Streptomyces globisporus
To Be Published
|
|
4HVM
 
 | | Crystal structure of tallysomycin biosynthesis protein TlmII | | Descriptor: | SULFATE ION, TlmII | | Authors: | Chang, C, Bigelow, L, Bearden, J, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-06 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Crystal structure of tallysomycin biosynthesis protein TlmII
To be Published
|
|
4K27
 
 | |
4I19
 
 | | The crystal structure of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus. | | Descriptor: | ACETATE ION, Epoxide hydrolase, FORMIC ACID | | Authors: | Tan, K, Bigelow, L, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-20 | | Release date: | 2012-12-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | The crystal structure of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus.
To be Published
|
|
4NEO
 
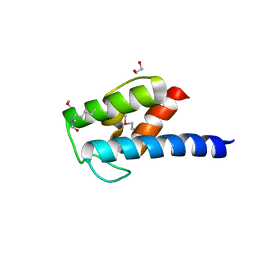 | | Structure of BlmI, a type-II acyl-carrier-protein from Streptomyces verticillus involved in bleomycin biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, Peptide synthetase NRPS type II-PCP | | Authors: | Cuff, M.E, Bigelow, L, Bearden, J, Babnigg, G, Bruno, C.J.P, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-10-29 | | Release date: | 2014-01-29 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of BlmI as a model for nonribosomal peptide synthetase peptidyl carrier proteins.
Proteins, 82, 2014
|
|
4I4J
 
 | | The structure of SgcE10, the ACP-polyene thioesterase involved in C-1027 biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, ACP-polyene thioesterase, D(-)-TARTARIC ACID, ... | | Authors: | Kim, Y, Bigelow, L, Bearden, J, Babnigg, J, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-27 | | Release date: | 2012-12-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.784 Å) | | Cite: | Crystal Structure of Thioesterase SgcE10 Supporting Common Polyene Intermediates in 9- and 10-Membered Enediyne Core Biosynthesis.
Acs Omega, 2, 2017
|
|
4IAG
 
 | | Crystal structure of ZbmA, the zorbamycin binding protein from Streptomyces flavoviridis | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Zbm binding protein | | Authors: | Cuff, M.E, Bigelow, L, Bruno, C.J.P, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-12-06 | | Release date: | 2013-02-20 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Zorbamycin-Binding Protein ZbmA, the Primary Self-Resistance Element in Streptomyces flavoviridis ATCC21892.
Biochemistry, 54, 2015
|
|
3I5N
 
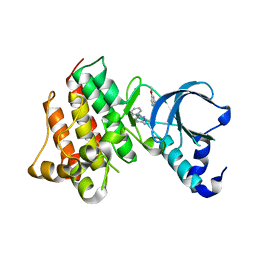 | | Crystal structure of c-Met with triazolopyridazine inhibitor 13 | | Descriptor: | 7-methoxy-N-[(6-phenyl[1,2,4]triazolo[4,3-b]pyridazin-3-yl)methyl]-1,5-naphthyridin-4-amine, Hepatocyte growth factor receptor | | Authors: | Bellon, S.F, Whittington, D.A, Long, A.M, Boezio, A.A. | | Deposit date: | 2009-07-06 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and optimization of potent and selective triazolopyridazine series of c-Met inhibitors
Bioorg.Med.Chem.Lett., 19, 2009
|
|
4FJY
 
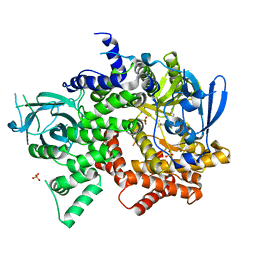 | | Crystal structure of PI3K-gamma in complex with quinoline-indoline inhibitor 24f | | Descriptor: | 4-[3,3-dimethyl-6-(morpholin-4-yl)-2,3-dihydro-1H-indol-1-yl]-7-fluoro-3-methyl-2-(pyridin-3-yl)quinoline, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2012-06-12 | | Release date: | 2012-10-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery and in Vivo Evaluation of Dual PI3K-beta/delta inhibitors
J.Med.Chem., 55, 2012
|
|
4FJZ
 
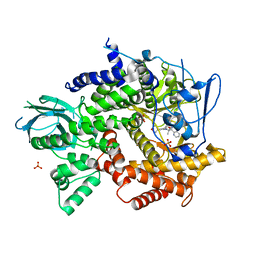 | | Crystal structure of PI3K-gamma in complex with pyrrolo-pyridine inhibitor 63 | | Descriptor: | 1'-[7-fluoro-3-methyl-2-(pyridin-2-yl)quinolin-4-yl]-6'-(morpholin-4-yl)-1',2,2',3,5,6-hexahydrospiro[pyran-4,3'-pyrrolo[3,2-b]pyridine], Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2012-06-12 | | Release date: | 2012-10-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery and in Vivo Evaluation of Dual PI3K-beta/delta inhibitors
J.Med.Chem., 55, 2012
|
|
5EYC
 
 | | Crystal structure of c-Met in complex with naphthyridinone inhibitor 5 | | Descriptor: | 6-[(1~{R})-1-[8-fluoranyl-6-(3-methyl-1,2-oxazol-5-yl)-[1,2,4]triazolo[4,3-a]pyridin-3-yl]ethyl]-1,6-naphthyridin-5-one, Hepatocyte growth factor receptor | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2015-11-24 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of (R)-6-(1-(8-Fluoro-6-(1-methyl-1H-pyrazol-4-yl)-[1,2,4]triazolo[4,3-a]pyridin-3-yl)ethyl)-3-(2-methoxyethoxy)-1,6-naphthyridin-5(6H)-one (AMG 337), a Potent and Selective Inhibitor of MET with High Unbound Target Coverage and Robust In Vivo Antitumor Activity.
J.Med.Chem., 59, 2016
|
|
5EYD
 
 | | Crystal structure of c-Met in complex with AMG 337 | | Descriptor: | 6-[(1~{R})-1-[8-fluoranyl-6-(1-methylpyrazol-4-yl)-[1,2,4]triazolo[4,3-a]pyridin-3-yl]ethyl]-3-(2-methoxyethoxy)-1,6-naphthyridin-5-one, Hepatocyte growth factor receptor | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2015-11-24 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of (R)-6-(1-(8-Fluoro-6-(1-methyl-1H-pyrazol-4-yl)-[1,2,4]triazolo[4,3-a]pyridin-3-yl)ethyl)-3-(2-methoxyethoxy)-1,6-naphthyridin-5(6H)-one (AMG 337), a Potent and Selective Inhibitor of MET with High Unbound Target Coverage and Robust In Vivo Antitumor Activity.
J.Med.Chem., 59, 2016
|
|
4XMO
 
 | | Crystal structure of c-Met in complex with (R)-5-(8-fluoro-3-(1-fluoro-1-(3-methoxyquinolin-6-yl)ethyl)-[1,2,4]triazolo[4,3-a]pyridin-6-yl)-3-methylisoxazole | | Descriptor: | 6-{(1R)-1-fluoro-1-[8-fluoro-6-(3-methyl-1,2-oxazol-5-yl)[1,2,4]triazolo[4,3-a]pyridin-3-yl]ethyl}-3-methoxyquinoline, Hepatocyte growth factor receptor | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2015-01-14 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of Potent and Selective 8-Fluorotriazolopyridine c-Met Inhibitors.
J.Med.Chem., 58, 2015
|
|
4XYF
 
 | | Crystal structure of c-Met in complex with (S)-5-(8-fluoro-3-(1-(3-(2-methoxyethoxy)quinolin-6-yl)ethyl)-[1,2,4]triazolo[4,3-a]pyridin-6-yl)-3-methylisoxazole | | Descriptor: | 6-{(1S)-1-[8-fluoro-6-(3-methyl-1,2-oxazol-5-yl)[1,2,4]triazolo[4,3-a]pyridin-3-yl]ethyl}-3-(2-methoxyethoxy)quinoline, Hepatocyte growth factor receptor | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2015-02-02 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of Potent and Selective 8-Fluorotriazolopyridine c-Met Inhibitors.
J.Med.Chem., 58, 2015
|
|
3CD8
 
 | | X-ray Structure of c-Met with triazolopyridazine Inhibitor. | | Descriptor: | 7-methoxy-4-[(6-phenyl[1,2,4]triazolo[4,3-b]pyridazin-3-yl)methoxy]quinoline, Hepatocyte growth factor receptor | | Authors: | Bellon, S.F, Albrecht, B.K, Harmange, J.-C, Bauer, D, Choquette, D, Dussault, I. | | Deposit date: | 2008-02-26 | | Release date: | 2008-04-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and Optimization of Triazolopyridazines as Potent and Selective Inhibitors of the c-Met Kinase.
J.Med.Chem., 51, 2008
|
|
3CCN
 
 | | X-ray structure of c-Met with triazolopyridazine inhibitor. | | Descriptor: | 4-[(6-phenyl[1,2,4]triazolo[4,3-b]pyridazin-3-yl)methyl]phenol, Hepatocyte growth factor receptor | | Authors: | Abrecht, B.K, Harmange, J.-C, Bauer, D, Dussault, I, long, A, Bellon, S.F. | | Deposit date: | 2008-02-26 | | Release date: | 2008-04-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Optimization of Triazolopyridazines as Potent and Selective Inhibitors of the c-Met Kinase.
J.Med.Chem., 51, 2008
|
|
4NNQ
 
 | | Crystal structure of LnmF protein from Streptomyces amphibiosporus | | Descriptor: | Putative enoyl-CoA hydratase, SULFATE ION | | Authors: | Michalska, K, Bigelow, L, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-11-18 | | Release date: | 2014-01-15 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: |
|
|
