4J2Q
 
 | |
5ZA0
 
 | |
4X98
 
 | | Immunoglobulin Fc heterodimer variant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-1 chain C region | | Authors: | Seok, S.H, Choi, H.J, Kim, Y.J, Seo, M.D, Kim, Y.S. | | Deposit date: | 2014-12-11 | | Release date: | 2015-06-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Crystal structures of immunoglobulin Fc heterodimers reveal the molecular basis for heterodimer formation.
Mol.Immunol., 65, 2015
|
|
4X99
 
 | | Immunoglobulin Fc heterodimers variant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-1 chain C region | | Authors: | Seok, S.H, Choi, H.J, Kim, Y.J, Seo, M.D, Kim, Y.S. | | Deposit date: | 2014-12-11 | | Release date: | 2015-06-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Crystal structures of immunoglobulin Fc heterodimers reveal the molecular basis for heterodimer formation.
Mol.Immunol., 65, 2015
|
|
3QD6
 
 | | Crystal structure of the CD40 and CD154 (CD40L) complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CD40 ligand, Tumor necrosis factor receptor superfamily member 5 | | Authors: | Lee, J.-O, Kim, Y.J, Song, D.H, Kim, H.M, Park, B.S. | | Deposit date: | 2011-01-18 | | Release date: | 2011-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystallographic and mutational analysis of the CD40-CD154 complex and its implications for receptor activation
J.Biol.Chem., 286, 2011
|
|
3DQB
 
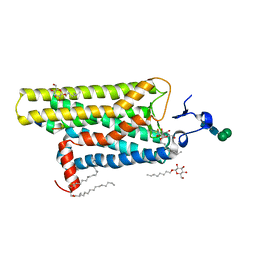 | | Crystal structure of the active G-protein-coupled receptor opsin in complex with a C-terminal peptide derived from the Galpha subunit of transducin | | Descriptor: | 11meric peptide form Guanine nucleotide-binding protein G(t) subunit alpha-1, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITIC ACID, ... | | Authors: | Scheerer, P, Park, J.H, Hildebrand, P.W, Kim, Y.J, Krauss, N, Choe, H.-W, Hofmann, K.P, Ernst, O.P. | | Deposit date: | 2008-07-09 | | Release date: | 2008-09-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of opsin in its G-protein-interacting conformation
Nature, 455, 2008
|
|
3WMK
 
 | |
4M4D
 
 | | Crystal structure of lipopolysaccharide binding protein | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Lipopolysaccharide-binding protein | | Authors: | Eckert, J.K, Kim, Y.J, Kim, J.I, Gurtler, K, Oh, D.Y, Ploeg, A.H, Pickkers, P, Lundvall, L, Hamann, L, Giamarellos-Bourboulis, E, Kubarenko, A.V, Weber, A.N, Kabesch, M, Kumpf, O, An, H.J, Lee, J.O, Schumann, R.R. | | Deposit date: | 2013-08-07 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.909 Å) | | Cite: | The crystal structure of lipopolysaccharide binding protein reveals the location of a frequent mutation that impairs innate immunity.
Immunity, 39, 2013
|
|
7VCJ
 
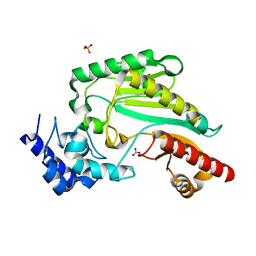 | | Arginine kinase H227A from Daphnia magna | | Descriptor: | Arginine kinase, NITRATE ION, PHOSPHATE ION | | Authors: | Kim, D.S, Jang, K, Kim, W.S, Kim, Y.J, Park, J.H. | | Deposit date: | 2021-09-03 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of H227A Mutant of Arginine Kinase in Daphnia magna Suggests the Importance of Its Stability.
Molecules, 27, 2022
|
|
7F1W
 
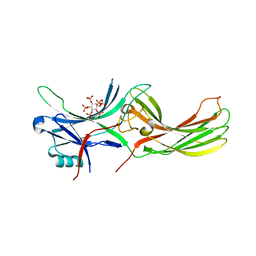 | | X-ray crystal structure of visual arrestin complexed with inositol hexaphosphate | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, S-arrestin | | Authors: | Kang, M, Jang, K, Eger, B.T, Ernst, O.P, Choe, H.W, Kim, Y.J. | | Deposit date: | 2021-06-10 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.097 Å) | | Cite: | Structural evidence for visual arrestin priming via complexation of phosphoinositols.
Structure, 30, 2022
|
|
7F1X
 
 | | X-ray crystal structure of visual arrestin complexed with inositol 1,4,5-triphosphate | | Descriptor: | 1,2-ETHANEDIOL, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, PENTANEDIAL, ... | | Authors: | Jang, K, Kang, M, Eger, B.T, Choe, H.W, Ernst, O.P, Kim, Y.J. | | Deposit date: | 2021-06-10 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural evidence for visual arrestin priming via complexation of phosphoinositols.
Structure, 30, 2022
|
|
3PQR
 
 | | Crystal structure of Metarhodopsin II in complex with a C-terminal peptide derived from the Galpha subunit of transducin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Guanine nucleotide-binding protein G(t) subunit alpha-1, ... | | Authors: | Choe, H.-W, Kim, Y.J, Park, J.H, Morizumi, T, Pai, E.F, Krauss, N, Hofmann, K.P, Scheerer, P, Ernst, O.P. | | Deposit date: | 2010-11-26 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of metarhodopsin II.
Nature, 471, 2011
|
|
3PXO
 
 | | Crystal structure of Metarhodopsin II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITIC ACID, RETINAL, ... | | Authors: | Choe, H.-W, Kim, Y.J, Park, J.H, Morizumi, T, Pai, E.F, Krauss, N, Hofmann, K.P, Scheerer, P, Ernst, O.P. | | Deposit date: | 2010-12-10 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of metarhodopsin II.
Nature, 471, 2011
|
|
4NHI
 
 | |
8PQO
 
 | | PITP in complex with the inhibitor VT01545 | | Descriptor: | (2~{R})-2-azanyl-3-[(2~{S},3~{R})-2-methyl-5-oxidanylidene-pyrrolidin-3-yl]sulfanyl-propanal, (2~{R})-3-methyl-2-(methylamino)butanoic acid, (2~{R})-4-methyl-2-(methylamino)pentanamide, ... | | Authors: | Boura, E, Eisenreichova, A. | | Deposit date: | 2023-07-11 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Non-vesicular phosphatidylinositol transfer plays critical roles in defining organelle lipid composition.
Embo J., 43, 2024
|
|
6C0V
 
 | |
8JZJ
 
 | | E.coli Glyceraldehyde-3-phosphate dehydrogenase structure under cryoprotect condition of ammonium sulfate | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, SULFATE ION | | Authors: | Jang, K, Hlaing, S.H.S, Kim, H.G, Kim, N, Choe, H.W, Kim, Y.J. | | Deposit date: | 2023-07-05 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | E.coli Glyceraldehyde-3-phosphate dehydrogenase structure under cryoprotect condition of ammonium sulfate
To Be Published
|
|
4TSV
 
 | |
6XFP
 
 | | Crystal Structure of BRAF kinase domain bound to Belvarafenib | | Descriptor: | 4-amino-N-{1-[(3-chloro-2-fluorophenyl)amino]-6-methylisoquinolin-5-yl}thieno[3,2-d]pyrimidine-7-carboxamide, CHLORIDE ION, Serine/threonine-protein kinase B-raf | | Authors: | Yin, J, Sudhamsu, J. | | Deposit date: | 2020-06-16 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ARAF mutations confer resistance to the RAF inhibitor belvarafenib in melanoma.
Nature, 594, 2021
|
|
7N9C
 
 | |
7N9E
 
 | |
7N9B
 
 | |
7N9T
 
 | |
8EZ6
 
 | |
5TSW
 
 | |
