225D
 
 | |
3UVT
 
 | |
6WUR
 
 | |
4Z2Z
 
 | |
5VXQ
 
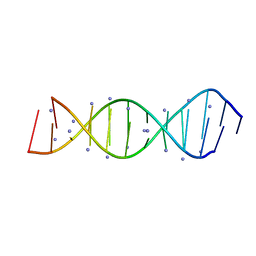 | | X-Ray crystallography structure of the parallel stranded duplex formed by 5-rA5-dA-rA5 | | Descriptor: | AMMONIUM ION, DNA/RNA (5'-R(*AP*AP*AP*AP*A)-D(P*A)-R(P*AP*AP*AP*AP*A)-3') | | Authors: | Xie, J, Chen, Y, Wei, X, Kozlov, G, Gehring, K. | | Deposit date: | 2017-05-23 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.002 Å) | | Cite: | Influence of nucleotide modifications at the C2' position on the Hoogsteen base-paired parallel-stranded duplex of poly(A) RNA.
Nucleic Acids Res., 45, 2017
|
|
5VMD
 
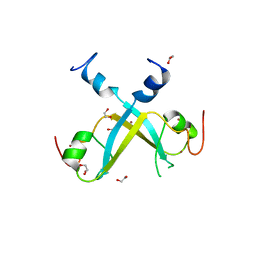 | | Crystal structure of UBR-box from UBR6 in a domain-swapping conformation | | Descriptor: | 1,2-ETHANEDIOL, F-box only protein 11, ZINC ION | | Authors: | Munoz-Escobar, J, Kozlov, G, Gehring, K. | | Deposit date: | 2017-04-27 | | Release date: | 2017-07-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Crystal structure of the UBR-box from UBR6/FBXO11 reveals domain swapping mediated by zinc binding.
Protein Sci., 26, 2017
|
|
8SMO
 
 | |
8CT8
 
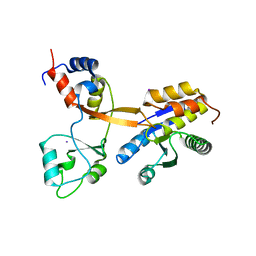 | | Crystal structure of Drosophila melanogaster PRL/CBS-pair domain complex | | Descriptor: | IODIDE ION, PRL-1 phosphatase, Unextended protein | | Authors: | Fakih, R, Goldstein, R.H, Kozlov, G, Gehring, K. | | Deposit date: | 2022-05-13 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Burst kinetics and CNNM binding are evolutionarily conserved properties of phosphatases of regenerating liver.
J.Biol.Chem., 299, 2023
|
|
1ME1
 
 | | Chimeric hairpin with 2',5'-linked RNA loop and RNA stem | | Descriptor: | 5'-R(*GP*GP*AP*CP*(U25)P*(U25)P*(C25)P*(5GP)P*GP*UP*CP*C)-3' | | Authors: | Denisov, A.Y, Hannoush, R.N, Gehring, K, Damha, M.J. | | Deposit date: | 2002-08-07 | | Release date: | 2003-08-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Novel RNA Motif Based on the Structure of Unusually Stable 2',5'-Linked r(UUCG) Loops
J.AM.CHEM.SOC., 125, 2003
|
|
1MK3
 
 | | SOLUTION STRUCTURE OF HUMAN BCL-W PROTEIN | | Descriptor: | Apoptosis regulator Bcl-W | | Authors: | Denisov, A.Y, Madiraju, M.S, Chen, G, Khadir, A, Beauparlant, P, Attardo, G, Shore, G.C, Gehring, K. | | Deposit date: | 2002-08-28 | | Release date: | 2003-06-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human BCL-w: modulation of ligand binding by the C-terminal helix
J.BIOL.CHEM., 278, 2003
|
|
6DJW
 
 | | Crystal Structure of pParkin (REP and RING2 deleted)-pUb-UbcH7 complex | | Descriptor: | RBR-type E3 ubiquitin transferase,RBR-type E3 ubiquitin transferase, Ubiquitin, Ubiquitin-conjugating enzyme E2 L3, ... | | Authors: | Sauve, V, Sung, G, Trempe, J.F, Gehring, K. | | Deposit date: | 2018-05-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.801 Å) | | Cite: | Mechanism of parkin activation by phosphorylation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DJ3
 
 | |
6PIK
 
 | | Tetrameric cryo-EM ArnA | | Descriptor: | Bifunctional polymyxin resistance protein ArnA, UDP-4-amino-4-deoxy-L-arabinose formyltransferase | | Authors: | Yang, M, Gehring, K. | | Deposit date: | 2019-06-26 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Cryo-electron microscopy structures of ArnA, a key enzyme for polymyxin resistance, revealed unexpected oligomerizations and domain movements.
J.Struct.Biol., 208, 2019
|
|
6PIH
 
 | | Hexameric ArnA cryo-EM structure | | Descriptor: | Bifunctional polymyxin resistance protein ArnA, UDP-4-amino-4-deoxy-L-arabinose formyltransferase | | Authors: | Yang, M, Gehring, K. | | Deposit date: | 2019-06-26 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Cryo-electron microscopy structures of ArnA, a key enzyme for polymyxin resistance, revealed unexpected oligomerizations and domain movements.
J.Struct.Biol., 208, 2019
|
|
6B3Y
 
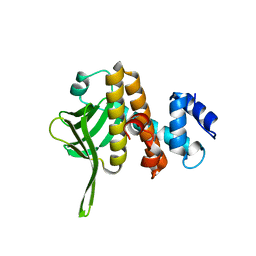 | | Crystal structure of the PH-like domain from DENND3 | | Descriptor: | DENN domain-containing protein 3 | | Authors: | Kozlov, G, Xu, J, Menade, M, Beaugrand, M, Pan, T, McPherson, P.S, Gehring, K. | | Deposit date: | 2017-09-25 | | Release date: | 2018-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | A PH-like domain of the Rab12 guanine nucleotide exchange factor DENND3 binds actin and is required for autophagy.
J. Biol. Chem., 293, 2018
|
|
8G91
 
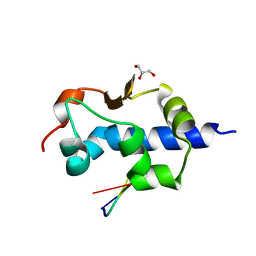 | |
8G90
 
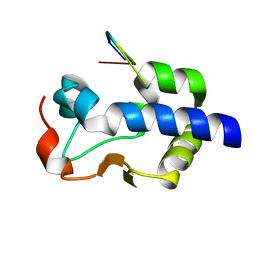 | |
3KUT
 
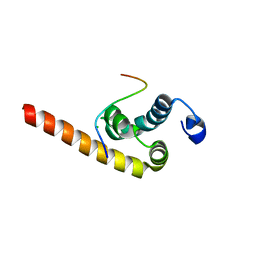 | |
3KUI
 
 | |
3KUR
 
 | |
3KUJ
 
 | |
3KUS
 
 | |
8GK6
 
 | |
4K95
 
 | | Crystal Structure of Parkin | | Descriptor: | E3 ubiquitin-protein ligase parkin, ZINC ION | | Authors: | Seirafi, M, Menade, M, Sauve, V, Kozlov, G, Trempe, J.-F, Nagar, B, Gehring, K. | | Deposit date: | 2013-04-19 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (6.499 Å) | | Cite: | Structure of parkin reveals mechanisms for ubiquitin ligase activation.
Science, 340, 2013
|
|
4K7D
 
 | | Crystal Structure of Parkin C-terminal RING domains | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase parkin, MALONATE ION, ... | | Authors: | Sauve, V, Trempe, J.-F, Menade, M, Gehring, K. | | Deposit date: | 2013-04-17 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of parkin reveals mechanisms for ubiquitin ligase activation.
Science, 340, 2013
|
|
