3G0X
 
 | | Human dihydroorotate dehydrogenase in complex with a leflunomide derivative inhibitor 5 | | Descriptor: | (2Z)-N-biphenyl-4-yl-2-cyano-3-cyclopropyl-3-hydroxyprop-2-enamide, Dihydroorotate dehydrogenase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Davies, M, Heikkila, T, McConkey, G.A, Fishwick, C.W.G, Parsons, M.R, Johnson, A.P. | | Deposit date: | 2009-01-29 | | Release date: | 2009-06-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based design, synthesis, and characterization of inhibitors of human and Plasmodium falciparum dihydroorotate dehydrogenases
J.Med.Chem., 52, 2009
|
|
7OZD
 
 | | FGFR1 kinase domain (residues 458-765) with mutations C488A, C584S in complex with 34. | | Descriptor: | 1,2-ETHANEDIOL, Fibroblast growth factor receptor 1, N-[6-(4-hydroxyphenyl)-1H-indazol-3-yl]benzamide, ... | | Authors: | Trinh, C.H, Turner, L.D, Fishwick, C.W.G. | | Deposit date: | 2021-06-27 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | From Fragment to Lead: De Novo Design and Development toward a Selective FGFR2 Inhibitor.
J.Med.Chem., 65, 2022
|
|
7OZB
 
 | | FGFR1 kinase domain (residues 458-765) with mutations C488A, C584S in complex with 38. | | Descriptor: | 1,2-ETHANEDIOL, 4-[3-(4-piperazin-4-ium-1-ylphenyl)-1H-indazol-6-yl]phenol, Fibroblast growth factor receptor 1, ... | | Authors: | Trinh, C.H, Turner, L.D, Fishwick, C.W.G. | | Deposit date: | 2021-06-27 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | From Fragment to Lead: De Novo Design and Development toward a Selective FGFR2 Inhibitor.
J.Med.Chem., 65, 2022
|
|
7OZF
 
 | | FGFR1 kinase domain (residues 458-765) with mutations C488A, C584S in complex with 19. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Fibroblast growth factor receptor 1, ... | | Authors: | Trinh, C.H, Turner, L.D, Fishwick, C.W.G. | | Deposit date: | 2021-06-28 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | From Fragment to Lead: De Novo Design and Development toward a Selective FGFR2 Inhibitor.
J.Med.Chem., 65, 2022
|
|
7OZY
 
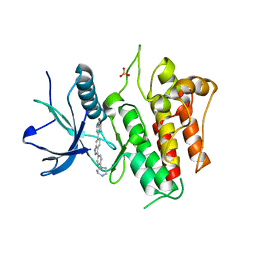 | | FGFR2 kinase domain (residues 461-763) in complex with 38. | | Descriptor: | 1,2-ETHANEDIOL, 4-[3-(4-piperazin-4-ium-1-ylphenyl)-1H-indazol-6-yl]phenol, Fibroblast growth factor receptor 2, ... | | Authors: | Trinh, C.H, Turner, L.D, Fishwick, C.W.G. | | Deposit date: | 2021-06-29 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | From Fragment to Lead: De Novo Design and Development toward a Selective FGFR2 Inhibitor.
J.Med.Chem., 65, 2022
|
|
3F1Q
 
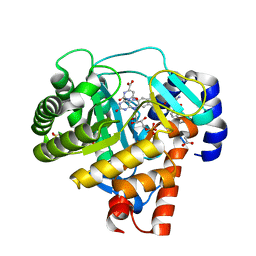 | | Human dihydroorotate dehydrogenase in complex with a leflunomide derivative inhibitor 1 | | Descriptor: | (2Z)-N-biphenyl-4-yl-2-cyano-3-hydroxybut-2-enamide, ACETIC ACID, Dihydroorotate dehydrogenase, ... | | Authors: | Heikkila, T, Davies, M, McConkey, G.A, Fishwick, C.W.G, Parsons, M.R, Johnson, A.P. | | Deposit date: | 2008-10-28 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design, synthesis, and characterization of inhibitors of human and Plasmodium falciparum dihydroorotate dehydrogenases
J.Med.Chem., 52, 2009
|
|
3G0U
 
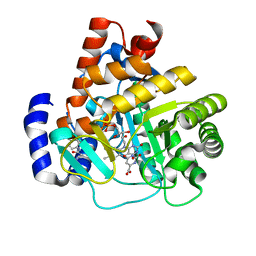 | | Human dihydroorotate dehydrogenase in complex with a leflunomide derivative inhibitor 4 | | Descriptor: | (2Z)-N-(3-chloro-2'-methoxybiphenyl-4-yl)-2-cyano-3-hydroxybut-2-enamide, Dihydroorotate dehydrogenase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Davis, M, Heikkila, T, McConkey, G.A, Fishwick, C.W.G, Parsons, M.R, Johnson, A.P. | | Deposit date: | 2009-01-28 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design, synthesis, and characterization of inhibitors of human and Plasmodium falciparum dihydroorotate dehydrogenases
J.Med.Chem., 52, 2009
|
|
4C5A
 
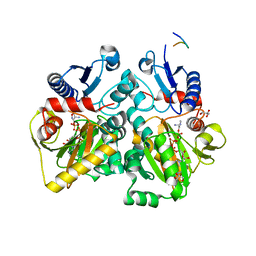 | | The X-ray crystal structures of D-alanyl-D-alanine ligase in complex ADP and D-cycloserine phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-ALANINE--D-ALANINE LIGASE, GLYCEROL, ... | | Authors: | Batson, S, Majce, V, Lloyd, A.J, Rea, D, Fishwick, C.W.G, Simmons, K.J, Fulop, V, Roper, D.I. | | Deposit date: | 2013-09-10 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Inhibition of D-Ala:D-Ala ligase through a phosphorylated form of the antibiotic D-cycloserine.
Nat Commun, 8, 2017
|
|
4C5B
 
 | | The X-ray crystal structure of D-alanyl-D-alanine ligase in complex with ATP and D-ala-D-ala | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CARBONATE ION, D-ALANINE, ... | | Authors: | Batson, S, Majce, V, Lloyd, A.J, Rea, D, Fishwick, C.W.G, Simmons, K.J, Fulop, V, Roper, D.I. | | Deposit date: | 2013-09-10 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Inhibition of D-Ala:D-Ala ligase through a phosphorylated form of the antibiotic D-cycloserine.
Nat Commun, 8, 2017
|
|
4C5C
 
 | | The X-ray crystal structure of D-alanyl-D-alanine ligase in complex with ADP and D-ala-D-ala | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-ALANINE, D-ALANINE--D-ALANINE LIGASE, ... | | Authors: | Batson, S, Majce, V, Lloyd, A.J, Rea, D, Fishwick, C.W.G, Simmons, K.J, Fulop, V, Roper, D.I. | | Deposit date: | 2013-09-10 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Inhibition of D-Ala:D-Ala ligase through a phosphorylated form of the antibiotic D-cycloserine.
Nat Commun, 8, 2017
|
|
5ML9
 
 | | Cocrystal structure of Fc gamma receptor IIIa interacting with Affimer F4, a specific binding protein which blocks IgG binding to the receptor. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Affimer F4 with specificity for Fc gamma receptor IIIa, CHLORIDE ION, ... | | Authors: | Robinson, J.I, Tomlinson, D.C, Baxter, E.W, Owen, R.L, Thomsen, M, Win, S.J, Nettleship, J.E, Tiede, C, Foster, R.J, Waterhouse, M.P, Harris, S.A, Owens, R.J, Fishwick, C.W.G, Goldman, A, McPherson, M.J, Morgan, A.W. | | Deposit date: | 2016-12-06 | | Release date: | 2017-12-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Affimer proteins inhibit immune complex binding to Fc gamma RIIIa with high specificity through competitive and allosteric modes of action.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6F8J
 
 | | Crystal Structure of E. coli GyraseB 24kDa in complex with 6-[(ethylcarbamoyl)amino]-4-(1H-pyrazol-1-yl)-N-(pyridin-3-yl)pyridine-3-carboxamide | | Descriptor: | 6-(ethylcarbamoylamino)-4-pyrazol-1-yl-~{N}-pyridin-3-yl-pyridine-3-carboxamide, DNA gyrase subunit B | | Authors: | Narramore, S.K, Stevenson, C.E.M, Lawson, D.M, Maxwell, A, Fishwick, C.W.G. | | Deposit date: | 2017-12-13 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | New insights into the binding mode of pyridine-3-carboxamide inhibitors of E. coli DNA gyrase.
Bioorg.Med.Chem., 27, 2019
|
|
6F96
 
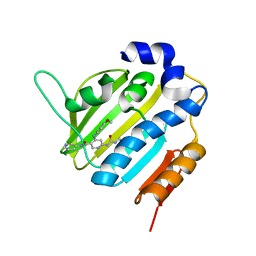 | | Crystal Structure of E. coli GyraseB 24kDa in complex with 6-[(ethylcarbamoyl)amino]-4-[(4-methoxyphenyl)amino]-N-(pyridin-3-yl)pyridine-3-carboxamide | | Descriptor: | 6-(ethylcarbamoylamino)-4-[(4-methoxyphenyl)amino]-~{N}-pyridin-3-yl-pyridine-3-carboxamide, DNA gyrase subunit B | | Authors: | Narramore, S.K, Stevenson, C.E.M, Lawson, D.M, Maxwell, A, Fishwick, C.W.G. | | Deposit date: | 2017-12-14 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | New insights into the binding mode of pyridine-3-carboxamide inhibitors of E. coli DNA gyrase.
Bioorg.Med.Chem., 27, 2019
|
|
6F86
 
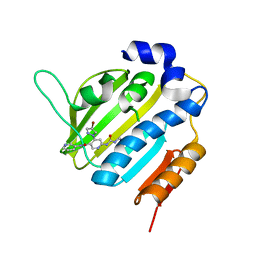 | | Crystal Structure of E. coli GyraseB 24kDa in complex with 4-(4-bromo-1H-pyrazol-1-yl)-6-[(ethylcarbamoyl)amino]-N-(pyridin-3-yl)pyridine-3-carboxamide | | Descriptor: | 4-(4-bromanylpyrazol-1-yl)-6-(ethylcarbamoylamino)-~{N}-pyridin-3-yl-pyridine-3-carboxamide, DNA gyrase subunit B | | Authors: | Narramore, S.K, Stevenson, C.E.M, Lawson, D.M, Maxwell, A, Fishwick, C.W.G. | | Deposit date: | 2017-12-12 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New insights into the binding mode of pyridine-3-carboxamide inhibitors of E. coli DNA gyrase.
Bioorg.Med.Chem., 27, 2019
|
|
6F94
 
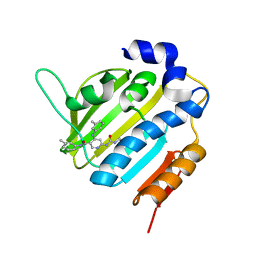 | | Crystal Structure of E. coli GyraseB 24kDa in complex with 6-[(ethylcarbamoyl)amino]-4-[(3-methyphenyl)amino]-N-(3-methyphenyl)pyridine-3-carboxamide | | Descriptor: | 6-(ethylcarbamoylamino)-~{N}-(3-methylphenyl)-4-[(3-methylphenyl)amino]pyridine-3-carboxamide, DNA gyrase subunit B | | Authors: | Narramore, S.K, Stevenson, C.E.M, Lawson, D.M, Maxwell, A, Fishwick, C.W.G. | | Deposit date: | 2017-12-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | New insights into the binding mode of pyridine-3-carboxamide inhibitors of E. coli DNA gyrase.
Bioorg.Med.Chem., 27, 2019
|
|
6FO6
 
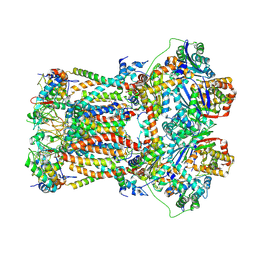 | | CryoEM structure of bovine cytochrome bc1 in complex with the anti-malarial inhibitor SCR0911 | | Descriptor: | 7-methoxy-3-methyl-2-[5-[4-(trifluoromethyloxy)phenyl]pyridin-3-yl]quinolin-4-ol, Cytochrome b, Cytochrome b-c1 complex subunit 1, ... | | Authors: | Johnson, R.M, Amporndanai, K, O'Neill, P.M, Fishwick, C.W.G, Jamson, A.H, Rawson, S.D, Hasnain, S.S, Antonyuk, S.V, Muench, S.P. | | Deposit date: | 2018-02-06 | | Release date: | 2018-02-28 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | X-ray and cryo-EM structures of inhibitor-bound cytochromebc1complexes for structure-based drug discovery.
IUCrJ, 5, 2018
|
|
6FO2
 
 | | CryoEM structure of bovine cytochrome bc1 with no ligand bound | | Descriptor: | Cytochrome b, Cytochrome b-c1 complex subunit 1, mitochondrial, ... | | Authors: | Johnson, R.M, Amporndanai, K, O'Neill, P.M, Fishwick, C.W.G, Jamson, A.H, Rawson, S.D, Hasnain, S.S, Antonyuk, S.V, Muench, S.P. | | Deposit date: | 2018-02-05 | | Release date: | 2018-02-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | X-ray and cryo-EM structures of inhibitor-bound cytochromebc1complexes for structure-based drug discovery.
IUCrJ, 5, 2018
|
|
6FO0
 
 | | CryoEM structure of bovine cytochrome bc1 in complex with the anti-malarial compound GSK932121 | | Descriptor: | 3-chloro-6-(hydroxymethyl)-2-methyl-5-{4-[3-(trifluoromethoxy)phenoxy]phenyl}pyridin-4-ol, Chain I/V, Cytochrome b, ... | | Authors: | Johnson, R.M, Amporndanai, K, O'Neill, P.M, Fishwick, C.W.G, Jamson, A.H, Rawson, S.D, Hasnain, S.S, Antonyuk, S.V, Muench, S.P. | | Deposit date: | 2018-02-05 | | Release date: | 2018-02-28 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | X-ray and cryo-EM structures of inhibitor-bound cytochromebc1complexes for structure-based drug discovery.
IUCrJ, 5, 2018
|
|
4KL1
 
 | | HCN4 CNBD in complex with cGMP | | Descriptor: | ACETATE ION, CYCLIC GUANOSINE MONOPHOSPHATE, GLYCEROL, ... | | Authors: | Lolicato, M, Arrigoni, C, Zucca, S, Nardini, M, Bucchi, A, Schroeder, I, Simmons, K, Bolognesi, M, DiFrancesco, D, Schwede, F, Fishwick, C.W.G, Johnson, A.P.K, Thiel, G, Moroni, A. | | Deposit date: | 2013-05-07 | | Release date: | 2014-04-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cyclic dinucleotides bind the C-linker of HCN4 to control channel cAMP responsiveness.
Nat.Chem.Biol., 10, 2014
|
|
5J8X
 
 | | CRYSTAL STRUCTURE OF E. COLI PBP5 WITH 2C | | Descriptor: | (4~{R})-4-[[4-(aminomethyl)phenyl]carbonylamino]-3,3-bis(oxidanyl)-2-oxa-3-boranuidabicyclo[4.4.0]deca-1(10),6,8-triene-10-carboxylic acid, D-alanyl-D-alanine carboxypeptidase DacA | | Authors: | Brem, J, Cain, R, McDonough, M.A, Clifton, I.J, Fishwick, C.W.G, Schofield, C.J. | | Deposit date: | 2016-04-08 | | Release date: | 2016-08-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural basis of metallo-beta-lactamase, serine-beta-lactamase and penicillin-binding protein inhibition by cyclic boronates.
Nat Commun, 7, 2016
|
|
5OKD
 
 | | Crystal structure of bovine Cytochrome bc1 in complex with inhibitor SCR0911. | | Descriptor: | 1,2-DIHEXANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | Authors: | Amporndanai, K, O'Neill, P.M, Hasnain, S.S, Antonyuk, S.V. | | Deposit date: | 2017-07-25 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | X-ray and cryo-EM structures of inhibitor-bound cytochromebc1complexes for structure-based drug discovery.
IUCrJ, 5, 2018
|
|
5BXI
 
 | | 1.7 Angstrom Resolution Crystal Structure of Putative Nucleoside Diphosphate Kinase from Toxoplasma gondii with Tyrosine of Tag Bound to Active Site | | Descriptor: | BICARBONATE ION, DI(HYDROXYETHYL)ETHER, Nucleoside diphosphate kinase | | Authors: | Minasov, G, Ruan, J, Ngo, H, Shuvalova, L, Dubrovska, I, Flores, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-06-09 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | CSGID Solves Structures and Identifies Phenotypes for Five Enzymes in Toxoplasma gondii .
Front Cell Infect Microbiol, 8, 2018
|
|
6I30
 
 | | Crystal structure of the AmpC from Pseudomonas aeruginosa with 1C | | Descriptor: | (3R)-3-(cyclohexylcarbonylamino)-2-oxidanyl-3,4-dihydro-1,2-benzoxaborinine-8-carboxylic acid, Class C beta-lactamase PDC-301 | | Authors: | Brem, J, Cahill, S.T, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2018-11-02 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Studies on the inhibition of AmpC and other beta-lactamases by cyclic boronates.
Biochim Biophys Acta Gen Subj, 1863, 2019
|
|
5FQ9
 
 | | Crystal structure of the OXA10 with 1C | | Descriptor: | (3R)-3-(cyclohexylcarbonylamino)-2-oxidanyl-3,4-dihydro-1,2-benzoxaborinine-8-carboxylic acid, ACETATE ION, BETA-LACTAMASE OXA-10, ... | | Authors: | Brem, J, McDonough, M.A, Cain, R, Clifton, I, Fishwick, C.W.G, Schofield, C.J. | | Deposit date: | 2015-12-08 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of metallo-beta-lactamase, serine-beta-lactamase and penicillin-binding protein inhibition by cyclic boronates.
Nat Commun, 7, 2016
|
|
5FQC
 
 | | Crystal structure of the metallo-beta-lactamase VIM-2 with 2C | | Descriptor: | (4~{R})-4-[[4-(aminomethyl)phenyl]carbonylamino]-3,3-bis(oxidanyl)-2-oxa-3-boranuidabicyclo[4.4.0]deca-1(10),6,8-triene-10-carboxylic acid, BETA-LACTAMASE, DIMETHYL SULFOXIDE, ... | | Authors: | Brem, J, Cain, R, McDonough, M.A, Clifton, I.J, Fishwick, C.W.G, Schofield, C.J. | | Deposit date: | 2015-12-08 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Structural basis of metallo-beta-lactamase, serine-beta-lactamase and penicillin-binding protein inhibition by cyclic boronates.
Nat Commun, 7, 2016
|
|
