2WIU
 
 | | Mercury-modified bacterial persistence regulator hipBA | | Descriptor: | CHLORIDE ION, HTH-TYPE TRANSCRIPTIONAL REGULATOR HIPB, MERCURY (II) ION, ... | | Authors: | Evdokimov, A, Voznesensky, I, Fennell, K, Anderson, M, Smith, J.F, Fisher, D.A. | | Deposit date: | 2009-05-17 | | Release date: | 2009-07-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | New Kinase Regulation Mechanism Found in Hipba: A Bacterial Persistence Switch.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3PES
 
 | | Crystal structure of uncharacterized protein from Pseudomonas phage YuA | | Descriptor: | Uncharacterized protein gp49 | | Authors: | Nocek, B, Stein, A, Evdokimove, A, Egorova, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-10-27 | | Release date: | 2010-12-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of uncharacterized protein from Pseudomonas phage YuA
TO BE PUBLISHED
|
|
7ML9
 
 | | The Mpp75Aa1.1 beta-pore-forming protein from Brevibacillus laterosporus | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Insecticidal protein, ... | | Authors: | Rydel, T.J, Zheng, M, Evdokimov, A. | | Deposit date: | 2021-04-27 | | Release date: | 2021-05-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural and functional characterization of Mpp75Aa1.1, a putative beta-pore forming protein from Brevibacillus laterosporus active against the western corn rootworm.
Plos One, 16, 2021
|
|
4RHZ
 
 | | Crystal structure of Cry23Aa1 and Cry37Aa1 binary protein complex | | Descriptor: | CALCIUM ION, Cry23AA1, Cry37AA1, ... | | Authors: | Rydel, T.J, Williams, J.M, Brown, G.R, Guzov, V.M, Sturman, E.J, Evdokimov, A. | | Deposit date: | 2014-10-03 | | Release date: | 2015-10-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Associated Bacillus thuringiensis Binary Protein Complex of Cry23Aa1 and Cry37Aa1: Crystal Structure, Insecticidal Data, and Pore Formation Modeling.
To be Published
|
|
4NG6
 
 | | The effects of Lysine 200 and Phenylalanine 239 Farnesyl Pyrophosphate Synthase (FPPS) mutations on the catalytic activity, crystal structure and inhibition by nitrogen containing bisphosphonates | | Descriptor: | 1-HYDROXY-2-(3-PYRIDINYL)ETHYLIDENE BIS-PHOSPHONIC ACID, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Farnesyl pyrophosphate synthase, ... | | Authors: | Tsoumpra, M.K, Muniz, J.R.C, Barnett, B.L, Pilka, E, Kwaasi, A.A, Kavanagh, K.L, Evdokimov, A, Walter, R.L, Ebetino, F.H, Oppermann, U, Russell, R.G.G, Dunford, J.E. | | Deposit date: | 2013-11-01 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The effects of Lysine 200 and Phenylalanine 239 Farnesyl Pyrophosphate Synthase (FPPS) mutations on the catalytic activity, crystal structure and inhibition by nitrogen containing bisphosphonates
TO BE PUBLISHED
|
|
4N9U
 
 | | The role of lysine 200 in the human farnesyl pyrophosphate synthase catalytic mechanism and the mode of inhibition by the nitrogen-containing bisphosphonates | | Descriptor: | 1,2-ETHANEDIOL, 1-HYDROXY-2-(3-PYRIDINYL)ETHYLIDENE BIS-PHOSPHONIC ACID, Farnesyl pyrophosphate synthase, ... | | Authors: | Tsoumpra, M.K, Muniz, J.R.C, Barnett, B.L, Pilka, E, Kwaasi, A.A, Kavanagh, K.L, Evdokimov, A, Walter, R.L, Ebetino, F.H, Oppermann, U, Russell, R.G.G, Dunford, J.E. | | Deposit date: | 2013-10-21 | | Release date: | 2014-10-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The role of lysine 200 in the human farnesyl pyrophosphate synthase catalytic mechanism and the mode of inhibition by the nitrogen-containing bisphosphonates
TO BE PUBLISHED
|
|
2QIS
 
 | | Crystal structure of human farnesyl pyrophosphate synthase T210S mutant bound to risedronate | | Descriptor: | 1-HYDROXY-2-(3-PYRIDINYL)ETHYLIDENE BIS-PHOSPHONIC ACID, Farnesyl pyrophosphate synthetase, MAGNESIUM ION | | Authors: | Kavanagh, K.L, Dunford, J.E, Hozjan, V, Evdokimov, A, Gileadi, O, von Delft, F, Weigelt, J, Arrowsmith, C.H, Sundstrom, M, Edwards, A, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-05 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human farnesyl pyrophosphate synthase T210S mutant bound to risedronate.
To be Published
|
|
4Q23
 
 | | The role of threonine 201 and tyrosine 204 in the human farnesyl pyrophosphate synthase catalytic mechanism and the mode of inhibition by the nitrogen-containing bisphosphonates | | Descriptor: | 1-HYDROXY-2-(3-PYRIDINYL)ETHYLIDENE BIS-PHOSPHONIC ACID, Farnesyl pyrophosphate synthase, MAGNESIUM ION, ... | | Authors: | Tsoumpra, M.K, Muniz, J.R.C, Barnett, B.L, Kwaasi, A.A, Pilka, E.S, Kavanagh, K.L, Evdokimov, A, Walter, R.L, Ebetino, F.H, von Delft, F, Oppermann, U, Russell, R.G.G, Dunford, J.E. | | Deposit date: | 2014-04-05 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The role of threonine 201 and tyrosine 204 in the human farnesyl pyrophosphate synthase catalytic mechanism and the mode of inhibition by the nitrogen-containing bisphosphonates
To be Published
|
|
3TCM
 
 | | Crystal Structure of Alanine Aminotransferase from Hordeum vulgare | | Descriptor: | Alanine aminotransferase 2, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE | | Authors: | Rydel, T.J, Sturman, E.J, Halls, C, Chen, S, Zeng, J, Evdokimov, A, Duff, S.M.G. | | Deposit date: | 2011-08-09 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | The Enzymology of alanine aminotransferase (AlaAT) isoforms from Hordeum vulgare and other organisms, and the HvAlaAT crystal structure.
Arch.Biochem.Biophys., 528, 2012
|
|
3UE8
 
 | | Kynurenine Aminotransferase II Inhibitors | | Descriptor: | (5-hydroxy-4-{[(1-hydroxy-2-oxo-1,2-dihydroquinolin-3-yl)amino]methyl}-6-methylpyridin-3-yl)methyl dihydrogen phosphate, CHLORIDE ION, Kynurenine/alpha-aminoadipate aminotransferase, ... | | Authors: | Dounay, A.B, Anderson, M, Bechle, B.M, Campbell, B.M, Claffey, M.M, Evdokimov, A, Edelweiss, E, Fonseca, K.R, Gan, X, Ghosh, S, Hayward, M.M, Horner, W, Kim, J.Y, McAllister, L.A, Pandit, J, Paradis, V, Parikh, V.D, Reese, M.R, Rong, S.B, Salafia, M.A, Schuyten, K, Strick, C.A, Tuttle, J.B, Valentine, J, Wang, H, Zawadzke, L.E, Verhoest, P.R. | | Deposit date: | 2011-10-28 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Discovery of Brain-Penetrant, Irreversible Kynurenine Aminotransferase II Inhibitors for Schizophrenia.
ACS Med Chem Lett, 3, 2012
|
|
3PBT
 
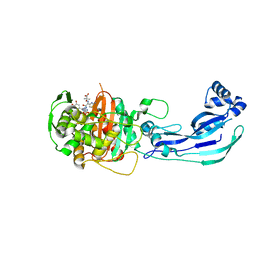 | | Crystal structure of PBP3 complexed with MC-1 | | Descriptor: | (4S,7Z)-7-(2-amino-1,3-thiazol-4-yl)-1-[({4-[(2R)-2,3-dihydroxypropyl]-3-(4,5-dihydroxypyridin-2-yl)-5-oxo-4,5-dihydro- 1H-1,2,4-triazol-1-yl}sulfonyl)amino]-4-formyl-10,10-dimethyl-1,6-dioxo-9-oxa-2,5,8-triazaundec-7-en-11-oate, Penicillin-binding protein 3 | | Authors: | Han, S, Evdokimov, A. | | Deposit date: | 2010-10-20 | | Release date: | 2010-12-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | Structural basis for effectiveness of siderophore-conjugated monocarbams against clinically relevant strains of Pseudomonas aeruginosa.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7S4E
 
 | |
6WPC
 
 | |
6OWK
 
 | |
2J9G
 
 | |
2C00
 
 | |
6D0O
 
 | | rdpA dioxygenase holoenzyme | | Descriptor: | (R)-phenoxypropionate/alpha-ketoglutarate-dioxygenase, 2-OXOGLUTARIC ACID, COBALT (II) ION | | Authors: | Rydel, T.J, Sturman, E.J, Zheng, M, Evdokimov, A. | | Deposit date: | 2018-04-10 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Development of enzymes for robust aryloxyphenoxypropionate and synthetic auxin herbicide tolerance traits in maize and soybean crops.
Pest Manag. Sci., 75, 2019
|
|
2VPQ
 
 | |
2VR1
 
 | |
2HK5
 
 | | Hck Kinase in Complex with Lck targetted Inhibitor PG-1009247 | | Descriptor: | 3-{[2-(1H-BENZIMIDAZOL-1-YL)-6-{[2-(DIETHYLAMINO)ETHYL]AMINO}PYRIMIDIN-4-YL]AMINO}-4-METHYLPHENOL, Tyrosine-protein kinase HCK | | Authors: | Walter, R.L, Mekel, M.J, Evdokimov, A.G, Pokross, M.E, Sabat, M.P. | | Deposit date: | 2006-07-03 | | Release date: | 2006-09-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The development of 2-benzimidazole substituted pyrimidine based inhibitors of lymphocyte specific kinase (Lck).
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2VQD
 
 | |
3TPE
 
 | | The phipa p3121 structure | | Descriptor: | Serine/threonine-protein kinase HipA | | Authors: | Schumacher, M.A, Link, T, Brennan, R.G. | | Deposit date: | 2011-09-07 | | Release date: | 2012-10-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Role of Unusual P Loop Ejection and Autophosphorylation in HipA-Mediated Persistence and Multidrug Tolerance.
Cell Rep, 2, 2012
|
|
3TPV
 
 | | Structure of pHipA bound to ADP | | Descriptor: | ADENINE, SULFATE ION, Serine/threonine-protein kinase HipA | | Authors: | Schumacher, M.A, Link, T.M, Brennan, R.G. | | Deposit date: | 2011-09-08 | | Release date: | 2012-10-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of Unusual P Loop Ejection and Autophosphorylation in HipA-Mediated Persistence and Multidrug Tolerance.
Cell Rep, 2, 2012
|
|
3TPT
 
 | | Structure of HipA(D309Q) bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | schumacher, M.A, link, T, Brennan, R.G. | | Deposit date: | 2011-09-08 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Role of Unusual P Loop Ejection and Autophosphorylation in HipA-Mediated Persistence and Multidrug Tolerance.
Cell Rep, 2, 2012
|
|
3TPD
 
 | | Structure of pHipA, monoclinic form | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Serine/threonine-protein kinase HipA | | Authors: | schumacher, M.A, link, T, Brennan, R.G. | | Deposit date: | 2011-09-07 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Role of Unusual P Loop Ejection and Autophosphorylation in HipA-Mediated Persistence and Multidrug Tolerance.
Cell Rep, 2, 2012
|
|
