2IFS
 
 | |
1A3L
 
 | |
1C1E
 
 | | CRYSTAL STRUCTURE OF A DIELS-ALDERASE CATALYTIC ANTIBODY 1E9 IN COMPLEX WITH ITS HAPTEN | | Descriptor: | 1,7,8,9,10,10-HEXACHLORO-4-METHYL-4-AZA-TRICYCLO[5.2.1.0(2,6)]DEC-8-ENE-3,5-DIONE, CATALYTIC ANTIBODY 1E9 (HEAVY CHAIN), CATALYTIC ANTIBODY 1E9 (LIGHT CHAIN), ... | | Authors: | Xu, J, Wilson, I.A. | | Deposit date: | 1999-07-22 | | Release date: | 2000-03-01 | | Last modified: | 2013-01-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution of shape complementarity and catalytic efficiency from a primordial antibody template.
Science, 286, 1999
|
|
7LQ1
 
 | | HUMAN PI3KDELTA IN COMPLEX WITH COMPOUND 28 | | Descriptor: | CHLORIDE ION, N-(5-(6-(2-((2S,6R)-2,6-dimethylmorpholino)pyridin-4-yl)-1-oxoisoindolin-4-yl)-2-methoxypyridin-3-yl)methanesulfonamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Lesburg, C.A, Augustin, M. | | Deposit date: | 2021-02-12 | | Release date: | 2022-02-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Discovery of a new series of PI3K-delta inhibitors from Virtual Screening.
Bioorg.Med.Chem.Lett., 42, 2021
|
|
1WUT
 
 | | Acyl Ureas as Human Liver Glycogen Phosphorylase Inhibitors for the Treatment of Type 2 Diabetes | | Descriptor: | 7-[2,6-DICHLORO-4-({[(2-CHLOROBENZOYL)AMINO]CARBONYL}AMINO)PHENOXY]HEPTANOIC ACID, Glycogen phosphorylase, muscle form, ... | | Authors: | Klabunde, T, Wendt, K.U, Kadereit, D, Brachvogel, V, Burger, H.-J, Herling, A.W, Oikonomakos, N.G, Kosmopoulou, M.N, Schmoll, D, Sarubbi, E, von Roedern, E, Schonafinger, K, Defossa, E. | | Deposit date: | 2004-12-08 | | Release date: | 2005-12-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystallographic studies on acyl ureas, a new class of glycogen phosphorylase inhibitors, as potential antidiabetic drugs
Protein Sci., 14, 2005
|
|
8SO9
 
 | | Phosphoinositide phosphate 3 kinase gamma | | Descriptor: | Phosphoinositide 3-kinase regulatory subunit 5, phosphatidylinositol-4,5-bisphosphate 3-kinase | | Authors: | Chen, C.-L, Tesmer, J.J.G. | | Deposit date: | 2023-04-28 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Molecular basis for G beta gamma-mediated activation of phosphoinositide 3-kinase gamma.
Nat.Struct.Mol.Biol., 2024
|
|
8SOA
 
 | | Phosphoinositide phosphate 3 kinase gamma bound with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Phosphoinositide 3-kinase regulatory subunit 5, phosphatidylinositol-4,5-bisphosphate 3-kinase | | Authors: | Chen, C.-L, Tesmer, J.J.G. | | Deposit date: | 2023-04-28 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Molecular basis for G beta gamma-mediated activation of phosphoinositide 3-kinase gamma.
Nat.Struct.Mol.Biol., 2024
|
|
8SOE
 
 | |
8SOD
 
 | |
8SOC
 
 | | Phosphoinositide phosphate 3 kinase gamma bound with ADP and Gbetagamma | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Chen, C.-L, Tesmer, J.J.G. | | Deposit date: | 2023-04-28 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis for G beta gamma-mediated activation of phosphoinositide 3-kinase gamma.
Nat.Struct.Mol.Biol., 2024
|
|
8SOB
 
 | | Phosphoinositide phosphate 3 kinase gamma bound with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Phosphoinositide 3-kinase regulatory subunit 5, phosphatidylinositol-4,5-bisphosphate 3-kinase | | Authors: | Chen, C.-L, Tesmer, J.J.G. | | Deposit date: | 2023-04-28 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular basis for G beta gamma-mediated activation of phosphoinositide 3-kinase gamma.
Nat.Struct.Mol.Biol., 2024
|
|
1RUR
 
 | | Crystal Structure (I) of native Diels-Alder antibody 13G5 Fab at pH 8.0 with a data set collected at SSRL beamline 9-1. | | Descriptor: | ZINC ION, immunoglobulin 13G5, heavy chain, ... | | Authors: | Zhu, X, Wentworth Jr, P, Wentworth, A.D, Eschenmoser, A, Lerner, R.A, Wilson, I.A. | | Deposit date: | 2003-12-11 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Probing the antibody-catalyzed water-oxidation pathway at atomic resolution.
Proc.Natl.Acad.Sci.USA, 110, 2004
|
|
1RUQ
 
 | | Crystal Structure (H) of u.v.-irradiated Diels-Alder antibody 13G5 Fab at pH 8.0 with a data set collected in house. | | Descriptor: | ZINC ION, immunoglobulin 13G5 heavy chain, immunoglobulin 13G5 light chain | | Authors: | Zhu, X, Wentworth Jr, P, Wentworth, A.D, Eschenmoser, A, Lerner, R.A, Wilson, I.A. | | Deposit date: | 2003-12-11 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Probing the antibody-catalyzed water-oxidation pathway at atomic resolution.
Proc.Natl.Acad.Sci.USA, 110, 2004
|
|
6AKW
 
 | | Crystal structure of RNA dioxygenase bound with an inhibitor | | Descriptor: | 2-OXOGLUTARIC ACID, 2-[[2,6-bis(chloranyl)-4-(3,5-dimethyl-1,2-oxazol-4-yl)phenyl]amino]benzoic acid, Alpha-ketoglutarate-dependent dioxygenase FTO | | Authors: | Yang, C.-G, Huang, Y, Gan, J. | | Deposit date: | 2018-09-04 | | Release date: | 2019-05-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Small-Molecule Targeting of Oncogenic FTO Demethylase in Acute Myeloid Leukemia.
Cancer Cell, 35, 2019
|
|
7WD0
 
 | | SARS-CoV-2 Beta spike in complex with two S5D2 Fabs | | Descriptor: | Heavy chain of S5D2 Fab, Light chain of S5D2 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-20 | | Release date: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
7WDF
 
 | | SARS-CoV-2 Beta spike in complex with two S3H3 Fabs | | Descriptor: | Heavy chain of S3H3 Fab, Light chain of S3H3 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-21 | | Release date: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
7WD9
 
 | |
7WD7
 
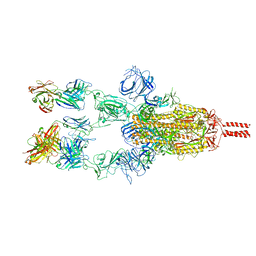 | |
7WCZ
 
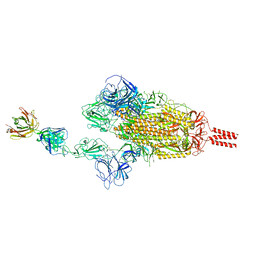 | | SARS-CoV-2 Beta spike in complex with one S5D2 Fab | | Descriptor: | Heavy chain of S5D2 Fab, Light chain of S5D2 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-20 | | Release date: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
7WCR
 
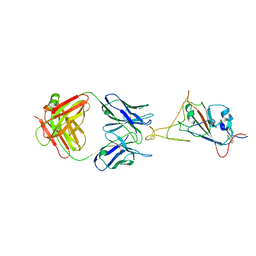 | |
7WD8
 
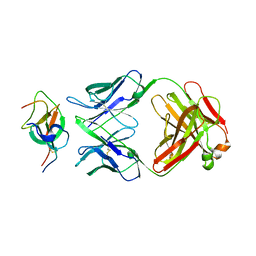 | | SARS-CoV-2 Beta spike SD1 in complex with S3H3 Fab | | Descriptor: | Heavy chain of S3H3 Fab, Light chain of S3H3 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-21 | | Release date: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
7TB1
 
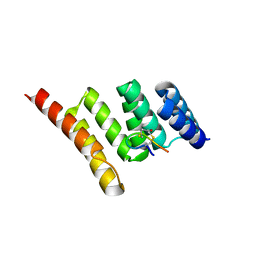 | | Crystal structure of STUB1 with a macrocyclic peptide | | Descriptor: | 1,3-bis(sulfanyl)propan-2-one, ALA-CYS-SER-SER-ILE-TRP-CYS-PRO-ASP-GLY, E3 ubiquitin-protein ligase CHIP | | Authors: | Bahmanjah, S, Klein, D.J. | | Deposit date: | 2021-12-21 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.785 Å) | | Cite: | Discovery and Structure-Based Design of Macrocyclic Peptides Targeting STUB1.
J.Med.Chem., 2022
|
|
7DCX
 
 | |
7DK6
 
 | |
7DD8
 
 | |
