8I48
 
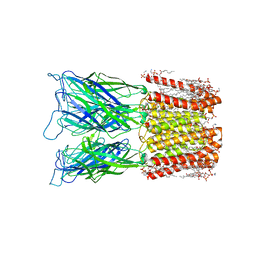 | | Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 4 in closed state | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHLORIDE ION, Proton-gated ion channel | | Authors: | Bharambe, N, Li, Z, Basak, S. | | Deposit date: | 2023-01-18 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Cryo-EM structures of prokaryotic ligand-gated ion channel GLIC provide insights into gating in a lipid environment.
Nat Commun, 15, 2024
|
|
8I47
 
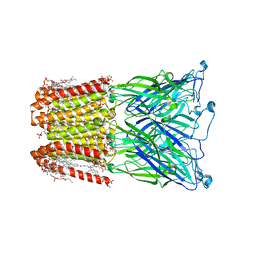 | |
5KGL
 
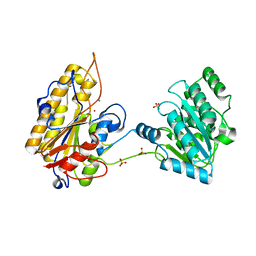 | | 2.45A resolution structure of Apo independent phosphoglycerate mutase from C. elegans (orthorhombic form) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Yu, H, Dranchak, P, MacArthur, R, Li, Z, Carlow, T, Suga, H, Inglese, J. | | Deposit date: | 2016-06-13 | | Release date: | 2017-04-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Macrocycle peptides delineate locked-open inhibition mechanism for microorganism phosphoglycerate mutases.
Nat Commun, 8, 2017
|
|
5JFI
 
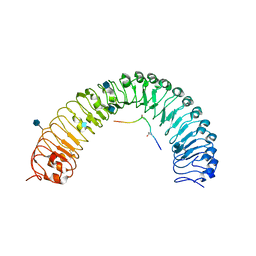 | | Crystal structure of a TDIF-TDR complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CLE41, Leucine-rich repeat receptor-like protein kinase TDR | | Authors: | Xu, G, Li, Z. | | Deposit date: | 2016-04-19 | | Release date: | 2017-03-29 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.749 Å) | | Cite: | Crystal structure of a TDIF-TDR complex
To Be Published
|
|
5DV9
 
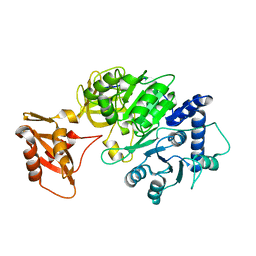 | |
3KUP
 
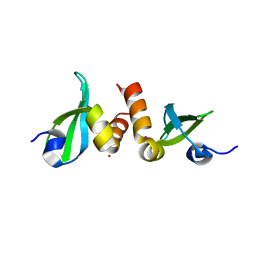 | | Crystal Structure of the CBX3 Chromo Shadow Domain | | Descriptor: | Chromobox protein homolog 3, UNKNOWN ATOM OR ION | | Authors: | Tempel, W, Li, Z, Li, Y, Kozieradzki, I, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-11-27 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal Structure of the CBX3 Chromo Shadow Domain
to be published
|
|
3C5W
 
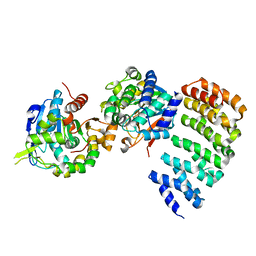 | | Complex between PP2A-specific methylesterase PME-1 and PP2A core enzyme | | Descriptor: | PP2A A subunit, PP2A C subunit, PP2A-specific methylesterase PME-1 | | Authors: | Xing, Y, Li, Z, Chen, Y, Stock, J, Jeffrey, P.D, Shi, Y. | | Deposit date: | 2008-02-01 | | Release date: | 2008-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural mechanism of demethylation and inactivation of protein phosphatase 2A.
Cell(Cambridge,Mass.), 133, 2008
|
|
3R93
 
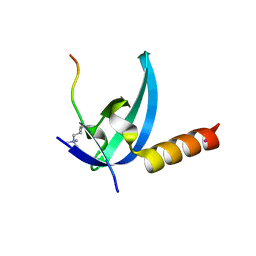 | | Crystal structure of the chromo domain of M-phase phosphoprotein 8 bound to H3K9Me3 peptide | | Descriptor: | H3K9Me3 peptide, M-phase phosphoprotein 8, UNKNOWN ATOM OR ION | | Authors: | Li, J, Li, Z, Ruan, J, Xu, C, Tong, Y, Pan, P.W, Tempel, W, Crombet, L, Min, J, Zang, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-03-24 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.057 Å) | | Cite: | Structural basis for specific binding of human MPP8 chromodomain to histone H3 methylated at lysine 9.
Plos One, 6, 2011
|
|
5DWV
 
 | | Crystal structure of the Luciferase complexed with substrate analogue | | Descriptor: | 2-[6-(cyclobuta-1,3-dien-1-ylamino)-1,3-benzothiazol-2-yl]-1,3-thiazol-4-ol, Luciferin 4-monooxygenase | | Authors: | Su, J, Li, Z, Yuan, Z, Gu, L. | | Deposit date: | 2015-09-23 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the Luciferase complexed with substrate analogue
To Be Published
|
|
3C5V
 
 | | PP2A-specific methylesterase apo form (PME) | | Descriptor: | Protein phosphatase methylesterase 1 | | Authors: | Xing, Y, Li, Z, Chen, Y, Stock, J, Jeffrey, P.D, Shi, Y. | | Deposit date: | 2008-02-01 | | Release date: | 2008-04-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural mechanism of demethylation and inactivation of protein phosphatase 2A.
Cell(Cambridge,Mass.), 133, 2008
|
|
3Q1J
 
 | | Crystal structure of tudor domain 1 of human PHD finger protein 20 | | Descriptor: | PHD finger protein 20, UNKNOWN ATOM OR ION | | Authors: | Tempel, W, Li, Z, Wernimont, A.K, Chao, X, Bian, C, Lam, R, Crombet, L, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-12-17 | | Release date: | 2011-02-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of the Tudor domains of human PHF20 reveal novel structural variations on the Royal Family of proteins.
Febs Lett., 586, 2012
|
|
7JZW
 
 | | Cryo-EM structure of CRISPR-Cas surveillance complex with AcrIF4 | | Descriptor: | CRISPR repeat sequence, CRISPR type I-F/YPEST-associated protein Csy1, CRISPR type I-F/YPEST-associated protein Csy2, ... | | Authors: | Chang, L, Li, Z, Gabel, C. | | Deposit date: | 2020-09-02 | | Release date: | 2020-12-30 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for inhibition of the type I-F CRISPR-Cas surveillance complex by AcrIF4, AcrIF7 and AcrIF14.
Nucleic Acids Res., 49, 2021
|
|
7JZX
 
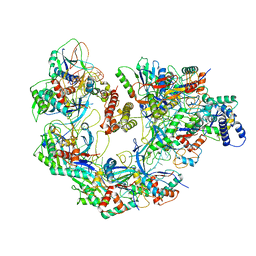 | | Cryo-EM structure of CRISPR-Cas surveillance complex with AcrIF7 | | Descriptor: | AcrF7, CRISPR type I-F/YPEST-associated protein Csy3, CRISPR-associated endonuclease Cas6/Csy4, ... | | Authors: | Chang, L, Li, Z, Gabel, C. | | Deposit date: | 2020-09-02 | | Release date: | 2020-12-30 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for inhibition of the type I-F CRISPR-Cas surveillance complex by AcrIF4, AcrIF7 and AcrIF14.
Nucleic Acids Res., 49, 2021
|
|
7JZZ
 
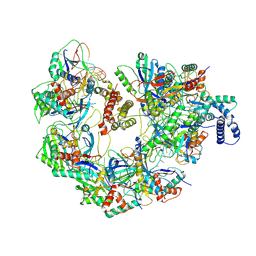 | | Cryo-EM structure of CRISPR-Cas surveillance complex with AcrIF14 | | Descriptor: | AcrF14, CRISPR type I-F/YPEST-associated protein Csy3, CRISPR-associated protein Csy1, ... | | Authors: | Chang, L, Li, Z, Gabel, C. | | Deposit date: | 2020-09-02 | | Release date: | 2020-12-30 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for inhibition of the type I-F CRISPR-Cas surveillance complex by AcrIF4, AcrIF7 and AcrIF14.
Nucleic Acids Res., 49, 2021
|
|
7JZY
 
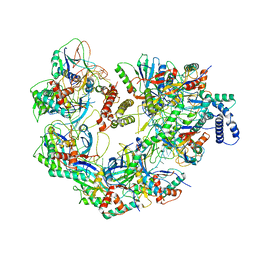 | | CryoEM structure of a CRISPR-Cas complex | | Descriptor: | AcrF9, CRISPR type I-F/YPEST-associated protein Csy3, CRISPR-associated protein Csy1, ... | | Authors: | Chang, L, Li, Z, Gabel, C. | | Deposit date: | 2020-09-02 | | Release date: | 2021-09-22 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | CryoEM structure of a CRISPR-Cas complex
To Be Published
|
|
4R05
 
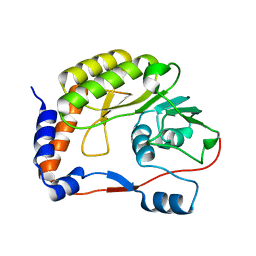 | | Crystal structure of the refolded DENV3 methyltransferase | | Descriptor: | Nonstructural protein NS5 | | Authors: | Brecher, M.B, Li, Z, Zhang, J, Chen, H, Lin, Q, Liu, B, Li, H.M. | | Deposit date: | 2014-07-29 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Refolding of a fully functional flavivirus methyltransferase revealed that S-adenosyl methionine but not S-adenosyl homocysteine is copurified with flavivirus methyltransferase.
Protein Sci., 24, 2015
|
|
1HW1
 
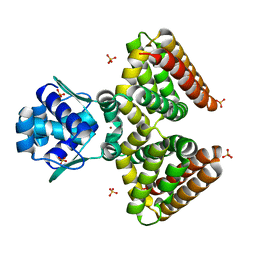 | | THE FADR-DNA COMPLEX: TRANSCRIPTIONAL CONTROL OF FATTY ACID METABOLISM IN ESCHERICHIA COLI | | Descriptor: | FATTY ACID METABOLISM REGULATOR PROTEIN, SULFATE ION, ZINC ION | | Authors: | Xu, Y, Heath, R.J, Li, Z, Rock, C.O, White, S.W. | | Deposit date: | 2001-01-09 | | Release date: | 2001-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The FadR.DNA complex. Transcriptional control of fatty acid metabolism in Escherichia coli.
J.Biol.Chem., 276, 2001
|
|
1HW2
 
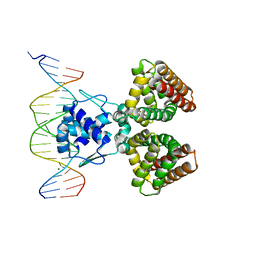 | | FADR-DNA COMPLEX: TRANSCRIPTIONAL CONTROL OF FATTY ACID METABOLISM IN ECHERICHIA COLI | | Descriptor: | 5'-D(*CP*GP*AP*TP*CP*TP*GP*GP*TP*CP*CP*GP*AP*CP*CP*AP*GP*AP*TP*GP*CP*T)-3', 5'-D(*G*CP*AP*TP*CP*TP*GP*GP*TP*CP*GP*GP*AP*CP*CP*AP*GP*AP*TP*CP*GP*A)-3', FATTY ACID METABOLISM REGULATOR PROTEIN, ... | | Authors: | Xu, Y, Heath, R.J, Li, Z, Rock, C.O, White, S.W. | | Deposit date: | 2001-01-09 | | Release date: | 2001-01-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The FadR.DNA complex. Transcriptional control of fatty acid metabolism in Escherichia coli.
J.Biol.Chem., 276, 2001
|
|
1M62
 
 | | Solution structure of the BAG domain from BAG4/SODD | | Descriptor: | BAG-family molecular chaperone regulator-4 | | Authors: | Briknarova, K, Takayama, S, Homma, S, Baker, K, Cabezas, E, Hoyt, D.W, Li, Z, Satterthwait, A.C, Ely, K.R. | | Deposit date: | 2002-07-11 | | Release date: | 2002-07-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | BAG4/SODD protein contains a short BAG domain.
J.Biol.Chem., 277, 2002
|
|
1DBU
 
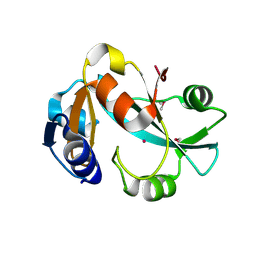 | | Crystal structure of cysteinyl-tRNA(Pro) deacylase protein from H. influenzae (HI1434) | | Descriptor: | MERCURY (II) ION, cysteinyl-tRNA(Pro) deacylase | | Authors: | Zhang, H, Huang, K, Li, Z, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 1999-11-03 | | Release date: | 2000-06-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of YbaK protein from Haemophilus influenzae (HI1434) at 1.8 A resolution: functional implications.
Proteins, 40, 2000
|
|
5H4P
 
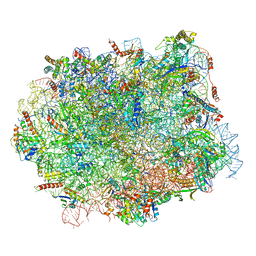 | | Structural snapshot of cytoplasmic pre-60S ribosomal particles bound with Nmd3, Lsg1, Tif6 and Reh1 | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Ma, C, Wu, S, Li, N, Chen, Y, Yan, K, Li, Z, Zheng, L, Lei, J, Woolford, J.L, Gao, N. | | Deposit date: | 2016-11-01 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural snapshot of cytoplasmic pre-60S ribosomal particles bound by Nmd3, Lsg1, Tif6 and Reh1
Nat. Struct. Mol. Biol., 24, 2017
|
|
1FJ8
 
 | | THE STRUCTURE OF BETA-KETOACYL-[ACYL CARRIER PROTEIN] SYNTHASE I IN COMPLEX WITH CERULENIN, IMPLICATIONS FOR DRUG DESIGN | | Descriptor: | (2S, 3R)-3-HYDROXY-4-OXO-7,10-TRANS,TRANS-DODECADIENAMIDE, BETA-KETOACYL-[ACYL CARRIER PROTEIN] SYNTHASE I | | Authors: | Price, A.C, Choi, K, Heath, R.J, Li, Z, White, S.W, Rock, C.O. | | Deposit date: | 2000-08-07 | | Release date: | 2000-08-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Inhibition of beta-ketoacyl-acyl carrier protein synthases by thiolactomycin and cerulenin. Structure and mechanism.
J.Biol.Chem., 276, 2001
|
|
3MTS
 
 | | Chromo Domain of Human Histone-Lysine N-Methyltransferase SUV39H1 | | Descriptor: | Histone-lysine N-methyltransferase SUV39H1 | | Authors: | Lam, R, Li, Z, Wang, J, Crombet, L, Walker, J.R, Ouyang, H, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-30 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Human SUV39H1 Chromodomain and Its Recognition of Histone H3K9me2/3.
Plos One, 7, 2012
|
|
4YOC
 
 | | Crystal Structure of human DNMT1 and USP7/HAUSP complex | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, Ubiquitin carboxyl-terminal hydrolase 7, ZINC ION | | Authors: | Cheng, J, Yang, H, Fang, J, Gong, R, Wang, P, Li, Z, Xu, Y. | | Deposit date: | 2015-03-11 | | Release date: | 2015-05-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.916 Å) | | Cite: | Molecular mechanism for USP7-mediated DNMT1 stabilization by acetylation.
Nat Commun, 6, 2015
|
|
1FJ4
 
 | | THE STRUCTURE OF BETA-KETOACYL-[ACYL CARRIER PROTEIN] SYNTHASE I IN COMPLEX WITH THIOLACTOMYCIN, IMPLICATIONS FOR DRUG DESIGN | | Descriptor: | BETA-KETOACYL-[ACYL CARRIER PROTEIN] SYNTHASE I, THIOLACTOMYCIN | | Authors: | Price, A.C, Choi, K, Heath, R.J, Li, Z, White, S.W, Rock, C.O. | | Deposit date: | 2000-08-07 | | Release date: | 2000-08-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Inhibition of beta-ketoacyl-acyl carrier protein synthases by thiolactomycin and cerulenin. Structure and mechanism.
J.Biol.Chem., 276, 2001
|
|
