7B2E
 
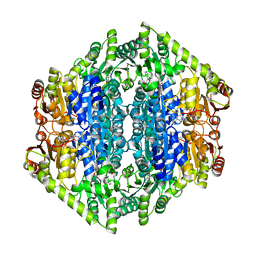 | | quadruple mutant of oxalyl-CoA decarboxylase from Methylorubrum extorquens with bound TPP and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative oxalyl-CoA decarboxylase (Oxc, ... | | Authors: | Pfister, P, Burgener, S, Nattermann, M, Zarzycki, J, Erb, T.J. | | Deposit date: | 2020-11-26 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Engineering a Highly Efficient Carboligase for Synthetic One-Carbon Metabolism.
Acs Catalysis, 11, 2021
|
|
7FD4
 
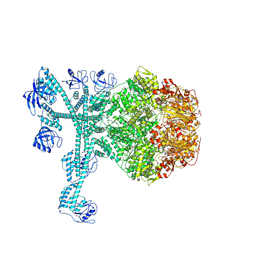 | | A complete three-dimensional structure of the Lon protease translocating a protein substrate (conformation 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Alpha-S1-casein, Lon protease, ... | | Authors: | Li, S, Hsieh, K, Kuo, C, Lee, S, Pintilie, G, Zhang, K, Chang, C. | | Deposit date: | 2021-07-16 | | Release date: | 2021-11-03 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Complete three-dimensional structures of the Lon protease translocating a protein substrate.
Sci Adv, 7, 2021
|
|
1MVE
 
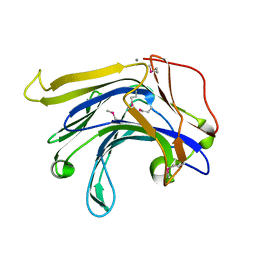 | | Crystal structure of a natural circularly-permutated jellyroll protein: 1,3-1,4-beta-D-glucanase from Fibrobacter succinogenes | | Descriptor: | CALCIUM ION, Truncated 1,3-1,4-beta-D-glucanase | | Authors: | Tsai, L.-C, Shyur, L.-F, Lee, S.-H, Lin, S.-S, Yuan, H.S. | | Deposit date: | 2002-09-25 | | Release date: | 2003-07-15 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of a Natural Circularly Permuted Jellyroll Protein: 1,3-1,4-beta-D-Glucanase from Fibrobacter succinogenes.
J.Mol.Biol., 330, 2003
|
|
7FD5
 
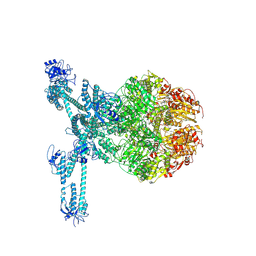 | | A complete three-dimensional structure of the Lon protease translocating a protein substrate (conformation 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Alpha-S1-casein, Lon protease, ... | | Authors: | Li, S, Hsieh, K, Kuo, C, Lee, S, Pintilie, G, Zhang, K, Chang, C. | | Deposit date: | 2021-07-16 | | Release date: | 2021-11-03 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Complete three-dimensional structures of the Lon protease translocating a protein substrate.
Sci Adv, 7, 2021
|
|
5D8D
 
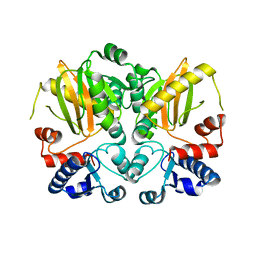 | |
5DMX
 
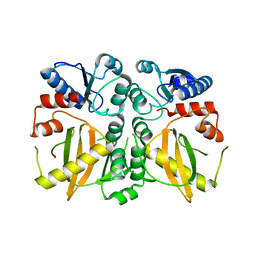 | |
3L6Y
 
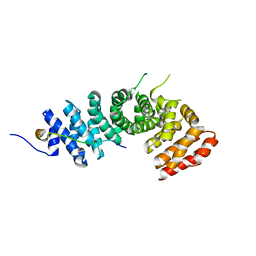 | | Crystal structure of p120 catenin in complex with E-cadherin | | Descriptor: | Catenin delta-1, E-cadherin | | Authors: | Ishiyama, N, Lee, S.-H, Liu, S, Li, G.-Y, Smith, M.J, Reichardt, L.F, Ikura, M. | | Deposit date: | 2009-12-27 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Dynamic and static interactions between p120 catenin and E-cadherin regulate the stability of cell-cell adhesion.
Cell(Cambridge,Mass.), 141, 2010
|
|
3L6X
 
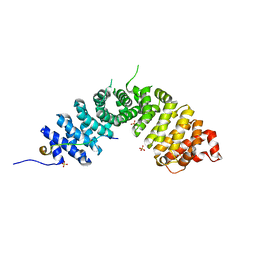 | | Crystal structure of p120 catenin in complex with E-cadherin | | Descriptor: | Catenin delta-1, E-cadherin, SULFATE ION | | Authors: | Ishiyama, N, Lee, S.-H, Liu, S, Li, G.-Y, Smith, M.J, Reichardt, L.F, Ikura, M. | | Deposit date: | 2009-12-27 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dynamic and static interactions between p120 catenin and E-cadherin regulate the stability of cell-cell adhesion.
Cell(Cambridge,Mass.), 141, 2010
|
|
2WZZ
 
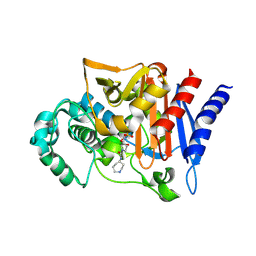 | |
4RX5
 
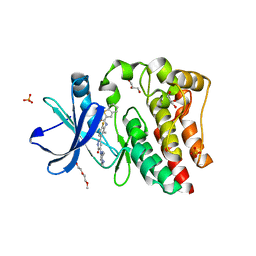 | | Bruton's tyrosine kinase (BTK) with pyridazinone compound 23 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, GLYCEROL, N-(6-fluoro-2-methyl-3-{5-[(5-methyl-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrazin-2-yl)amino]-6-oxo-1,6-dihydropyridazin-3-yl}phenyl)-1-benzothiophene-2-carboxamide, ... | | Authors: | Eigenbrot, C, Yu, C. | | Deposit date: | 2014-12-08 | | Release date: | 2015-12-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.356 Å) | | Cite: | Discovery of highly potent and selective Bruton's tyrosine kinase inhibitors: Pyridazinone analogs with improved metabolic stability.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
8AFV
 
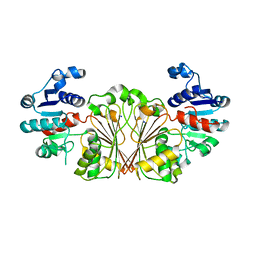 | | DaArgC3 - Engineered Formyl Phosphate Reductase with 3 substitutions (S178V, G182V, L233I) | | Descriptor: | N-acetyl-gamma-glutamyl-phosphate reductase, SODIUM ION | | Authors: | Pfister, P, Nattermann, M, Zarzycki, J, Erb, T.J. | | Deposit date: | 2022-07-18 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Engineering a new-to-nature cascade for phosphate-dependent formate to formaldehyde conversion in vitro and in vivo.
Nat Commun, 14, 2023
|
|
8AFU
 
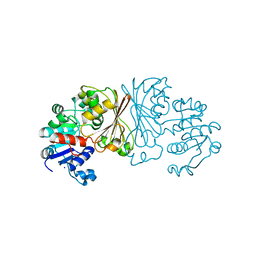 | | DaArgC - N-acetyl-gamma-glutamyl-phosphate Reductase of Denitrovibrio acetiphilus | | Descriptor: | N-acetyl-gamma-glutamyl-phosphate reductase, SODIUM ION | | Authors: | Pfister, P, Nattermann, M, Zarzycki, J, Erb, T.J. | | Deposit date: | 2022-07-18 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Engineering a new-to-nature cascade for phosphate-dependent formate to formaldehyde conversion in vitro and in vivo.
Nat Commun, 14, 2023
|
|
4DXD
 
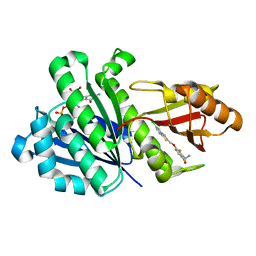 | | Staphylococcal Aureus FtsZ in complex with 723 | | Descriptor: | 3-[(6-chloro[1,3]thiazolo[5,4-b]pyridin-2-yl)methoxy]-2,6-difluorobenzamide, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Lu, J, Soisson, S.M. | | Deposit date: | 2012-02-27 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Restoring methicillin-resistant Staphylococcus aureus susceptibility to beta-lactam antibiotics.
Sci Transl Med, 4, 2012
|
|
5JJA
 
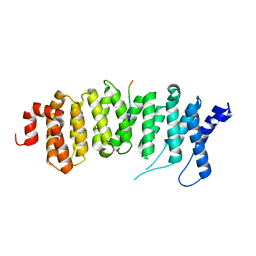 | | Crystal structure of a PP2A B56gamma/BubR1 complex | | Descriptor: | Mitotic checkpoint serine/threonine-protein kinase BUB1 beta, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform | | Authors: | Wang, Z, Wang, J, Rao, Z, Xu, W. | | Deposit date: | 2016-04-22 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of a PP2A B56-BubR1 complex and its implications for PP2A substrate recruitment and localization.
Protein Cell, 7, 2016
|
|
2WZX
 
 | |
5Y9J
 
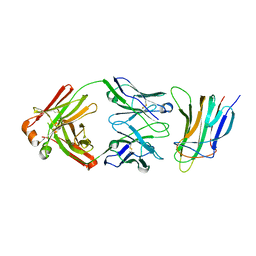 | | BAFF in complex with belimumab | | Descriptor: | Tumor necrosis factor ligand superfamily member 13B, belibumab light chain, belimumab heavy chain | | Authors: | Heo, Y.-S, Shin, W. | | Deposit date: | 2017-08-25 | | Release date: | 2018-02-21 | | Last modified: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | BAFF-neutralizing interaction of belimumab related to its therapeutic efficacy for treating systemic lupus erythematosus.
Nat Commun, 9, 2018
|
|
5Y9K
 
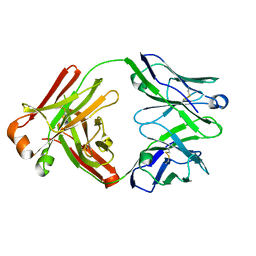 | | Structure of the belimumab Fab fragment | | Descriptor: | belimumab heavy chain, belimumab light chain | | Authors: | Heo, Y.-S, Shin, W. | | Deposit date: | 2017-08-25 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | BAFF-neutralizing interaction of belimumab related to its therapeutic efficacy for treating systemic lupus erythematosus.
Nat Commun, 9, 2018
|
|
8IKR
 
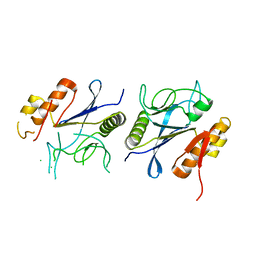 | | Crystal structure of DpaA | | Descriptor: | YkuD domain-containing protein | | Authors: | Wang, H.-J, Hsieh, K.-Y, Lee, S.-H, Chang, C.-I. | | Deposit date: | 2023-03-01 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the hydrolytic activity of the transpeptidase-like protein DpaA to detach Braun's lipoprotein from peptidoglycan.
Mbio, 14, 2023
|
|
4QD4
 
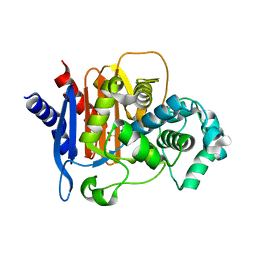 | | Structure of ADC-68, a Novel Carbapenem-Hydrolyzing Class C Extended-Spectrum -Lactamase from Acinetobacter baumannii | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-lactamase ADC-68, CITRIC ACID | | Authors: | Hong, M.K, Kang, L.W. | | Deposit date: | 2014-05-13 | | Release date: | 2015-01-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of ADC-68, a novel carbapenem-hydrolyzing class C extended-spectrum beta-lactamase isolated from Acinetobacter baumannii
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7WI1
 
 | |
5X8M
 
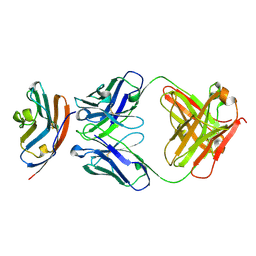 | | PD-L1 in complex with durvalumab | | Descriptor: | Programmed cell death 1 ligand 1, durvalumab heavy chain, durvalumab light chain | | Authors: | Heo, Y.S, Lee, H.T. | | Deposit date: | 2017-03-03 | | Release date: | 2017-08-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.661 Å) | | Cite: | Molecular mechanism of PD-1/PD-L1 blockade via anti-PD-L1 antibodies atezolizumab and durvalumab
Sci Rep, 7, 2017
|
|
5X8L
 
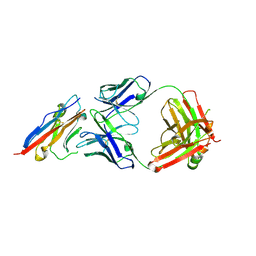 | | PD-L1 in complex with atezolizumab | | Descriptor: | Programmed cell death 1 ligand 1, atezolizumab heavy chain, atezolizumab light chain | | Authors: | Heo, Y.S, Lee, H.T. | | Deposit date: | 2017-03-03 | | Release date: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular mechanism of PD-1/PD-L1 blockade via anti-PD-L1 antibodies atezolizumab and durvalumab
Sci Rep, 7, 2017
|
|
3V6A
 
 | |
6JC2
 
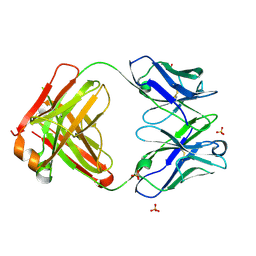 | | Crystal structure of the Fab fragment of ipilimumab | | Descriptor: | SULFATE ION, ipilimumab fab heavy chain, ipilimumab fab light chain | | Authors: | Heo, Y.S. | | Deposit date: | 2019-01-27 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structure of the Fab Fragment of an Anti-CTLA-4 Antibody, Ipilimumab, Used for Cancer Immunotherapy
Bull.Korean Chem.Soc., 40, 2019
|
|
6J52
 
 | | Crystal structure of CARD-only protein in frog virus 3 | | Descriptor: | Caspase recruitment domain-only protein | | Authors: | Park, H.H, Kwon, S. | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Structural transformation-mediated dimerization of caspase recruitment domain revealed by the crystal structure of CARD-only protein in frog virus 3.
J. Struct. Biol., 205, 2019
|
|
