4G0F
 
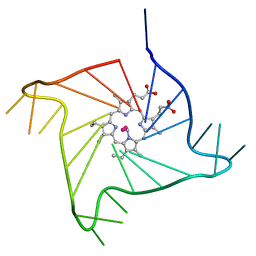 | | Crystal structure of the complex of a human telomeric repeat G-quadruplex and N-methyl mesoporphyrin IX (P6) | | Descriptor: | DNA (5'-D(*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*G)-3'), N-METHYLMESOPORPHYRIN, POTASSIUM ION | | Authors: | Nicoludis, J.M, Miller, S.T, Jeffrey, P, Lawton, T.J, Rosenzweig, A.C, Yatsunyk, L.A. | | Deposit date: | 2012-07-09 | | Release date: | 2012-12-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Optimized End-Stacking Provides Specificity of N-Methyl Mesoporphyrin IX for Human Telomeric G-Quadruplex DNA.
J.Am.Chem.Soc., 134, 2012
|
|
4FXM
 
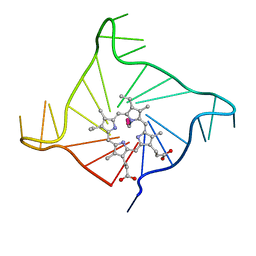 | | Crystal structure of the complex of a human telomeric repeat G-quadruplex and N-methyl mesoporphyrin IX (P21212) | | Descriptor: | DNA (5'-D(*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*G)-3'), N-METHYLMESOPORPHYRIN, POTASSIUM ION | | Authors: | Nicoludis, J.M, Miller, S.T, Jeffrey, P, Lawton, T.J, Rosenzweig, A.C, Yatsunyk, L.A. | | Deposit date: | 2012-07-03 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Optimized End-Stacking Provides Specificity of N-Methyl Mesoporphyrin IX for Human Telomeric G-Quadruplex DNA.
J.Am.Chem.Soc., 134, 2012
|
|
1PIL
 
 | |
4I5J
 
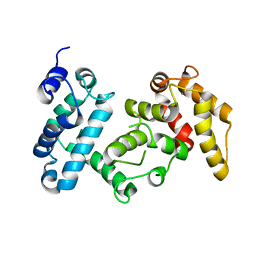 | | PP2A PR70 Holoenzyme | | Descriptor: | CALCIUM ION, Serine/threonine-protein phosphatase 2A regulatory subunit B'' subunit alpha | | Authors: | Xing, Y, Jeffery, P.D, Shi, Y. | | Deposit date: | 2012-11-28 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | Structure of the Ca(2+)-dependent PP2A heterotrimer and insights into Cdc6 dephosphorylation.
Cell Res., 23, 2013
|
|
6PRH
 
 | |
3IEI
 
 | |
4M9Z
 
 | | Crystal structure of CED-4 bound CED-3 fragment | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CED-3 fragment, Cell death protein 4, ... | | Authors: | Huang, W.J, Jinag, T.Y, Choi, W.Y, Wang, J.W, Shi, Y.G. | | Deposit date: | 2013-08-15 | | Release date: | 2013-10-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.405 Å) | | Cite: | Mechanistic insights into CED-4-mediated activation of CED-3.
Genes Dev., 27, 2013
|
|
4I5L
 
 | | Structural mechanism of trimeric PP2A holoenzyme involving PR70: insight for Cdc6 dephosphorylation | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, MALONATE ION, ... | | Authors: | Wlodarchak, N, Satyshur, K.A, Guo, F, Xing, Y. | | Deposit date: | 2012-11-28 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structure of the Ca(2+)-dependent PP2A heterotrimer and insights into Cdc6 dephosphorylation.
Cell Res., 23, 2013
|
|
4M9S
 
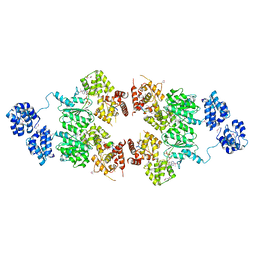 | | crystal structure of CED-4 bound CED-3 fragment | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CED-3 fragment, Cell death protein 4, ... | | Authors: | Huang, W.J, Jinag, T.Y, Choi, W.Y, Wang, J.W, Shi, Y.G. | | Deposit date: | 2013-08-15 | | Release date: | 2013-10-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.207 Å) | | Cite: | Mechanistic insights into CED-4-mediated activation of CED-3.
Genes Dev., 27, 2013
|
|
4M9X
 
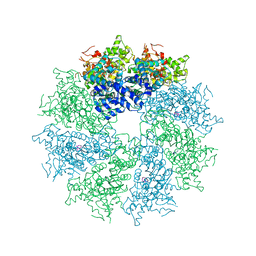 | | Crystal structure of CED-4 bound CED-3 fragment | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CED-3 fragment, Cell death protein 4, ... | | Authors: | Huang, W.J, Jinag, T.Y, Choi, W.Y, Wang, J.W, Shi, Y.G. | | Deposit date: | 2013-08-15 | | Release date: | 2013-10-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.344 Å) | | Cite: | Mechanistic insights into CED-4-mediated activation of CED-3.
Genes Dev., 27, 2013
|
|
6Z7C
 
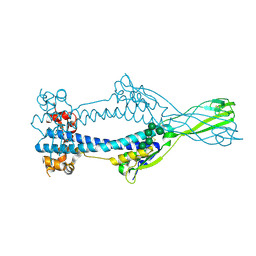 | | Variant Surface Glycoprotein VSGsur mutant H122A | | Descriptor: | Variant surface glycoprotein Sur, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Zeelen, J.P, Straaten van, M, Stebbins, C.E. | | Deposit date: | 2020-05-30 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure of trypanosome coat protein VSGsur and function in suramin resistance.
Nat Microbiol, 6, 2021
|
|
6Z7D
 
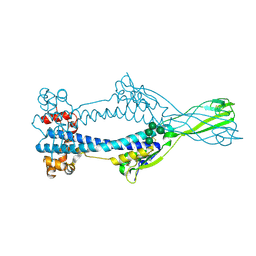 | | Variant Surface Glycoprotein VSGsur mutant H122A soaked in 0.77 mM Suramin. | | Descriptor: | Variant surface glycoprotein Sur, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Zeelen, J.P, Straaten van, M, Stebbins, C.E. | | Deposit date: | 2020-05-30 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of trypanosome coat protein VSGsur and function in suramin resistance.
Nat Microbiol, 6, 2021
|
|
6Z79
 
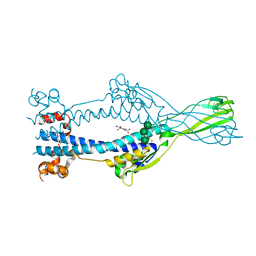 | | Variant Surface Glycoprotein VSGsur, I3C ("Magic Triangle") derivative used for phasing of the structure and subsequently as a model for molecular replacement of native, mutants, and drug soaks. | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, Variant surface glycoprotein Sur, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Zeelen, J.P, Straaten van, M, Stebbins, C.E. | | Deposit date: | 2020-05-30 | | Release date: | 2021-03-17 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of trypanosome coat protein VSGsur and function in suramin resistance.
Nat Microbiol, 6, 2021
|
|
6Z7E
 
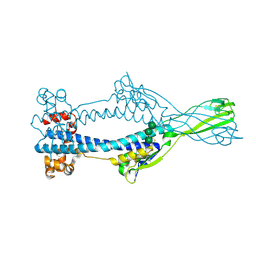 | |
6Z8G
 
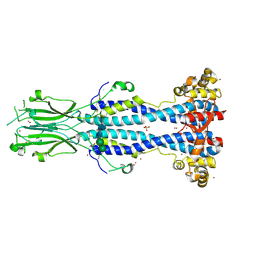 | | Crystal structure of VSG13 soaked in 0.5 M used to phase VSG13 to solve the structure. | | Descriptor: | BROMIDE ION, SULFATE ION, Variant surface glycoprotein MITat 1.13, ... | | Authors: | Stebbins, C.E, Hempelmann, A, Van Straaten, M, Zeelen, J. | | Deposit date: | 2020-06-02 | | Release date: | 2021-03-17 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure of trypanosome coat protein VSGsur and function in suramin resistance.
Nat Microbiol, 6, 2021
|
|
6Z8H
 
 | | Crystal structure of Variant Surface Glycoprotein VSG13 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Variant surface glycoprotein MITat 1.13, ... | | Authors: | Stebbins, C.E, Hempelmann, A, Van Straaten, M, Zeelen, J. | | Deposit date: | 2020-06-02 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structure of trypanosome coat protein VSGsur and function in suramin resistance.
Nat Microbiol, 6, 2021
|
|
6Z7A
 
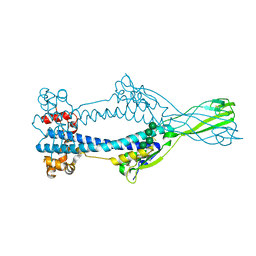 | | Variant Surface Glycoprotein VSGsur | | Descriptor: | Variant surface glycoprotein Sur, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Zeelen, J.P, Straaten van, M, Stebbins, C.E, Hashemi, H. | | Deposit date: | 2020-05-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | A Parasite Coat Protein Binds Suramin to Confer Drug Resistance
Nat Microbiol, 6, 2021
|
|
3P71
 
 | | Crystal structure of the complex of LCMT-1 and PP2A | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl](ethyl)amino}-5'-deoxyadenosine, DI(HYDROXYETHYL)ETHER, Leucine carboxyl methyltransferase 1, ... | | Authors: | Xing, Y, Stanevich, V, Satyshur, K.A, Jiang, L. | | Deposit date: | 2010-10-11 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Structural Basis for Tight Control of PP2A Methylation and Function by LCMT-1.
Mol.Cell, 41, 2011
|
|
1UCB
 
 | |
6UFF
 
 | |
6MWW
 
 | | LasR LBD:BB0126 complex | | Descriptor: | 4-[3-(methylsulfonyl)phenoxy]-N-[(1R,3R,5R)-2-oxobicyclo[3.1.0]hexan-3-yl]butanamide, Transcriptional regulator LasR | | Authors: | Bassler, B.L, Paczkowski, J.E. | | Deposit date: | 2018-10-30 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | An Autoinducer Analogue Reveals an Alternative Mode of Ligand Binding for the LasR Quorum-Sensing Receptor.
Acs Chem.Biol., 14, 2019
|
|
6MWL
 
 | | LasR LBD:mBTL complex | | Descriptor: | 4-(3-bromophenoxy)-N-[(3S)-2-oxothiolan-3-yl]butanamide, Transcriptional regulator LasR | | Authors: | Bassler, B.L, Paczkowski, J.E. | | Deposit date: | 2018-10-29 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | An Autoinducer Analogue Reveals an Alternative Mode of Ligand Binding for the LasR Quorum-Sensing Receptor.
Acs Chem.Biol., 14, 2019
|
|
6MWZ
 
 | | LasR LBD T75V/Y93F/A127W:BB0126 | | Descriptor: | 4-[3-(methylsulfonyl)phenoxy]-N-[(1S,3S,5S)-2-oxobicyclo[3.1.0]hexan-3-yl]butanamide, ALA-HIS-HIS-HIS-HIS-ALA, Transcriptional regulator LasR | | Authors: | Bassler, B.L, Paczkowski, J.E. | | Deposit date: | 2018-10-30 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.657 Å) | | Cite: | An Autoinducer Analogue Reveals an Alternative Mode of Ligand Binding for the LasR Quorum-Sensing Receptor.
Acs Chem.Biol., 14, 2019
|
|
6MVM
 
 | | LasR LBD L130F:3OC14HSL complex | | Descriptor: | 3-oxo-N-[(3S)-2-oxooxolan-3-yl]tetradecanamide, Transcriptional regulator LasR | | Authors: | Paczkowski, J.E, Bassler, B.L. | | Deposit date: | 2018-10-26 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | An Autoinducer Analogue Reveals an Alternative Mode of Ligand Binding for the LasR Quorum-Sensing Receptor.
Acs Chem.Biol., 14, 2019
|
|
6MWH
 
 | | LasR LBD:BB0020 complex | | Descriptor: | 2-(3-bromophenoxy)-N-[(1S,2S,3R,5S)-2-hydroxybicyclo[3.1.0]hexan-3-yl]acetamide, Transcriptional regulator LasR | | Authors: | Bassler, B.L, Paczkowski, J.E. | | Deposit date: | 2018-10-29 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | An Autoinducer Analogue Reveals an Alternative Mode of Ligand Binding for the LasR Quorum-Sensing Receptor.
Acs Chem.Biol., 14, 2019
|
|
