3Q5H
 
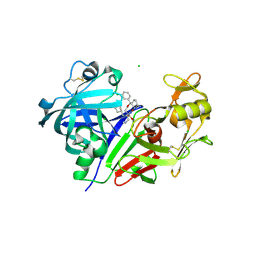 | | Clinically Useful Alkyl Amine Renin Inhibitors | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Renin, ... | | Authors: | Wu, Z, McKeever, B.M. | | Deposit date: | 2010-12-28 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Discovery of VTP-27999, an Alkyl Amine Renin Inhibitor with Potential for Clinical Utility.
ACS Med Chem Lett, 2, 2011
|
|
2LQZ
 
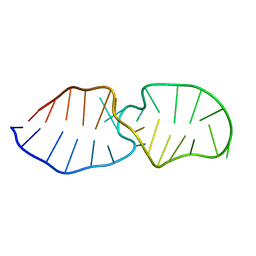 | |
5HXW
 
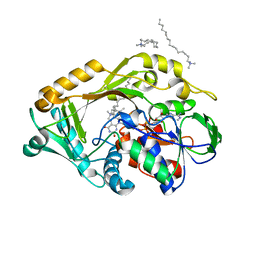 | | L-amino acid deaminase from Proteus vulgaris | | Descriptor: | CETYL-TRIMETHYL-AMMONIUM, FLAVIN-ADENINE DINUCLEOTIDE, L-amino acid deaminase | | Authors: | Zhou, H, Ju, Y, Niu, L, Teng, M. | | Deposit date: | 2016-01-31 | | Release date: | 2016-08-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Crystal structure of a membrane-bound l-amino acid deaminase from Proteus vulgaris
J.Struct.Biol., 195, 2016
|
|
5I39
 
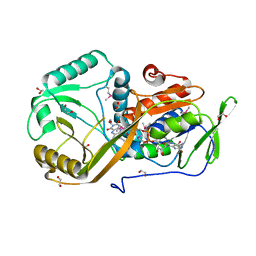 | | High resolution structure of L-amino acid deaminase from Proteus vulgaris with the deletion of the specific insertion sequence | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, L-amino acid deaminase | | Authors: | Zhou, H, Ju, Y, Niu, L, Teng, M. | | Deposit date: | 2016-02-10 | | Release date: | 2016-08-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of a membrane-bound l-amino acid deaminase from Proteus vulgaris
J.Struct.Biol., 195, 2016
|
|
4L58
 
 | |
7JZV
 
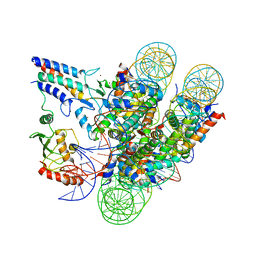 | | Cryo-EM structure of the BRCA1-UbcH5c/BARD1 E3-E2 module bound to a nucleosome | | Descriptor: | BRCA1,Ubiquitin-conjugating enzyme E2 D3, BRCA1-associated RING domain protein 1, Histone H2A type 2-A, ... | | Authors: | Witus, S.R, Burrell, A.L, Hansen, J.M, Farrell, D.P, Dimaio, F, Kollman, J.M, Klevit, R.E. | | Deposit date: | 2020-09-02 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | BRCA1/BARD1 site-specific ubiquitylation of nucleosomal H2A is directed by BARD1.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7CKW
 
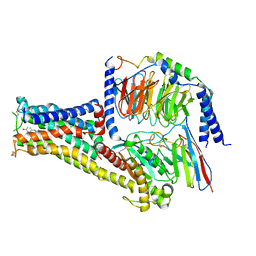 | | Cryo-EM structure of Fenoldopam bound dopamine receptor DRD1-Gs signaling complex | | Descriptor: | (1R)-6-chloranyl-1-(4-hydroxyphenyl)-2,3,4,5-tetrahydro-1H-3-benzazepine-7,8-diol, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Yan, W, Shao, W. | | Deposit date: | 2020-07-20 | | Release date: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Ligand recognition and allosteric regulation of DRD1-Gs signaling complexes.
Cell, 184, 2021
|
|
7VNG
 
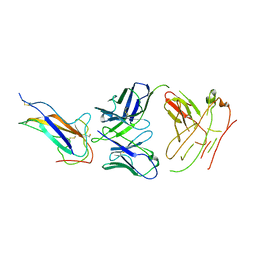 | | Crystal structure of human coronavirus 229E spike protein receptor-binding domain in complex with S11 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S11 Fab heavy chain, S11 Fab light chain, ... | | Authors: | Xiang, J.C, Zhao, W.W, Yang, B. | | Deposit date: | 2021-10-11 | | Release date: | 2022-10-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Antigenic mapping reveals sites of vulnerability on alpha-HCoV spike protein.
Commun Biol, 5, 2022
|
|
5GRS
 
 | | Complex structure of the fission yeast SREBP-SCAP binding domains | | Descriptor: | Sterol regulatory element-binding protein 1, Sterol regulatory element-binding protein cleavage-activating protein | | Authors: | Gong, X, Qian, H.W, Wu, J.P, Yan, N. | | Deposit date: | 2016-08-12 | | Release date: | 2016-12-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Complex structure of the fission yeast SREBP-SCAP binding domains reveals an oligomeric organization
Cell Res., 26, 2016
|
|
5GPD
 
 | |
7DAT
 
 | | The crystal structure of COVID-19 main protease treated by AF | | Descriptor: | COVID-19 MAIN PROTEASE, GOLD ION | | Authors: | He, Z.S, He, B, Cao, P, Jiang, H.D, Gong, Y, Gao, X.Y. | | Deposit date: | 2020-10-18 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A comparison of Remdesivir versus gold cluster in COVID-19 animal model: A better therapeutic outcome of gold cluster.
Nano Today, 44, 2022
|
|
7DAV
 
 | | The native crystal structure of COVID-19 main protease | | Descriptor: | COVID-19 MAIN PROTEASE | | Authors: | He, Z.S, He, B, Cao, P, Jiang, H.D, Gong, Y, Gao, X.Y. | | Deposit date: | 2020-10-18 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A comparison of Remdesivir versus gold cluster in COVID-19 animal model: A better therapeutic outcome of gold cluster.
Nano Today, 44, 2022
|
|
7DAU
 
 | | The crystal structure of COVID-19 main protease treated by GA | | Descriptor: | COVID-19 MAIN PROTEASE, GOLD ION | | Authors: | He, Z.S, He, B, Cao, P, Jiang, H.D, Gong, Y, Gao, X.Y. | | Deposit date: | 2020-10-18 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A comparison of Remdesivir versus gold cluster in COVID-19 animal model: A better therapeutic outcome of gold cluster.
Nano Today, 44, 2022
|
|
6JUI
 
 | | The atypical Myb-like protein Cdc5 contains two distinct nucleic acid-binding surfaces | | Descriptor: | Pre-mRNA-splicing factor CEF1 | | Authors: | Wang, C, Li, G, Li, M, Yang, J, Liu, J. | | Deposit date: | 2019-04-14 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Two distinct nucleic acid binding surfaces of Cdc5 regulate development.
Biochem.J., 476, 2019
|
|
6KF5
 
 | | Microbial Hormone-sensitive lipase E53 mutant I256L | | Descriptor: | (4-nitrophenyl) hexanoate, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Yang, X, Li, Z.Y, Li, J, Xu, X.W. | | Deposit date: | 2019-07-06 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Microbial Hormone-sensitive lipase E53 mutant I256L
To Be Published
|
|
6KI0
 
 | | Crystal Structure of Human ASC-CARD | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Apoptosis-associated speck-like protein containing a CARD, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Xu, Z.H, Jin, T.C. | | Deposit date: | 2019-07-16 | | Release date: | 2020-07-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Homotypic CARD-CARD interaction is critical for the activation of NLRP1 inflammasome.
Cell Death Dis, 12, 2021
|
|
6KF1
 
 | | Microbial Hormone-sensitive lipase- E53 mutant S162A | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, HEXANOIC ACID, ... | | Authors: | Xiaochen, Y, Zhengyang, L, Xuewei, X, Jixi, L. | | Deposit date: | 2019-07-05 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Functional and Structural Insights into Environmental Adaptation of a Novel Hormone-sensitive Lipase, E53, Obtained from Erythrobacter longus
To Be Published
|
|
6KSF
 
 | | Crystal Structure of ALKBH1 bound to 21-mer DNA bulge | | Descriptor: | Alpha-ketoglutarate-dependent dioxygenase alkB homolog 1, CHLORIDE ION, DNA (5'-D(*DGP*DCP*DTP*DGP*DAP*DGP*DTP*DGP*DCP*DCP*DCP*DGP*DCP*DGP*DTP*DGP*DCP*DTP*DGP*DGP*DAP*DTP*DCP*DC)-3'), ... | | Authors: | Li, H, Zhang, M. | | Deposit date: | 2019-08-23 | | Release date: | 2020-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mammalian ALKBH1 serves as an N6-mA demethylase of unpairing DNA.
Cell Res., 30, 2020
|
|
6KEU
 
 | | Wildtype E53, a microbial HSL esterase | | Descriptor: | (4-nitrophenyl) hexanoate, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Yang, X.C, Li, Z.Y, Xu, X.W, Li, J.X. | | Deposit date: | 2019-07-05 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Wildtype E53, a microbial HSL esterase
To Be Published
|
|
3W28
 
 | | The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E251A mutant with xylotriose | | Descriptor: | Glycoside hydrolase family 10, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-xylopyranose | | Authors: | Han, X, Gao, J, Shang, N, Huang, C.-H, Ko, T.-P, Zhu, Z, Wiegel, J, Shao, W, Guo, R.-T. | | Deposit date: | 2012-11-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural and functional analyses of catalytic domain of GH10 xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Proteins, 81, 2013
|
|
8IFP
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 1 | | Descriptor: | (1R,2S,5S)-3-[(2S)-2-(tert-butylcarbamoylamino)-3,3-dimethyl-butanoyl]-6,6-dimethyl-N-[(2S)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IFT
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 10 | | Descriptor: | (8S)-N-[(1S)-1-cyano-2-[(3S)-2-oxidanylidenepyrrolidin-3-yl]ethyl]-7-[(2S)-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-1,4-dithia-7-azaspiro[4.4]nonane-8-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IFQ
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 2 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IFS
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 7 | | Descriptor: | (8~{S})-7-[(2~{S})-2-(~{tert}-butylcarbamoylamino)-3,3-dimethyl-butanoyl]-~{N}-[(1~{S})-1-cyano-2-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]ethyl]-1,4-dithia-7-azaspiro[4.4]nonane-8-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
8IFR
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with compound 3 | | Descriptor: | (1R,2S,5S)-3-[(2S)-2-(tert-butylcarbamoylamino)-3,3-dimethyl-butanoyl]-6,6-dimethyl-N-[(2S)-5-oxidanylidene-1-[(3S)-2-oxidanylidenepyrrolidin-3-yl]hex-3-en-2-yl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Nie, T.Q, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-02-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir.
Nat Commun, 14, 2023
|
|
