7USE
 
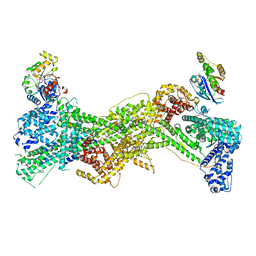 | | Cryo-EM structure of WAVE regulatory complex with Rac1 bound on both A and D site | | Descriptor: | Abl interactor 2, Cytoplasmic FMR1-interacting protein 1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Ding, B, Yang, S, Chen, B, Chowdhury, S. | | Deposit date: | 2022-04-25 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures reveal a key mechanism of WAVE regulatory complex activation by Rac1 GTPase.
Nat Commun, 13, 2022
|
|
7USC
 
 | | Cryo-EM structure of WAVE Regulatory Complex | | Descriptor: | Abl interactor 2, Cytoplasmic FMR1-interacting protein 1, Nck-associated protein 1, ... | | Authors: | Ding, B, Yang, S, Chen, B, Chowdhury, S. | | Deposit date: | 2022-04-25 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures reveal a key mechanism of WAVE regulatory complex activation by Rac1 GTPase.
Nat Commun, 13, 2022
|
|
7USD
 
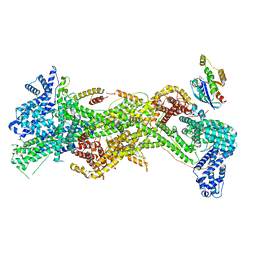 | | Cryo-EM structure of D-site Rac1-bound WAVE Regulatory Complex | | Descriptor: | Abl interactor 2, Cytoplasmic FMR1-interacting protein 1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Ding, B, Yang, S, Chen, B, Chowdhury, S. | | Deposit date: | 2022-04-25 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures reveal a key mechanism of WAVE regulatory complex activation by Rac1 GTPase.
Nat Commun, 13, 2022
|
|
7D88
 
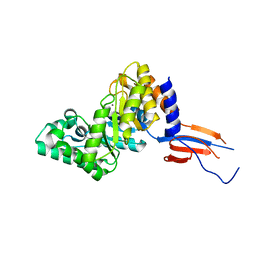 | |
7D89
 
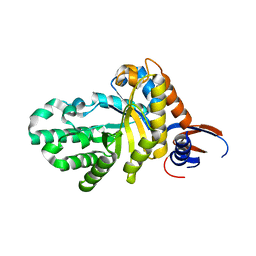 | |
1QFW
 
 | | TERNARY COMPLEX OF HUMAN CHORIONIC GONADOTROPIN WITH FV ANTI ALPHA SUBUNIT AND FV ANTI BETA SUBUNIT | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY (ANTI ALPHA SUBUNIT) (HEAVY CHAIN), ANTIBODY (ANTI ALPHA SUBUNIT) (LIGHT CHAIN), ... | | Authors: | Tegoni, M, Spinelli, S, Cambillau, C. | | Deposit date: | 1999-04-15 | | Release date: | 2000-04-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of a ternary complex between human chorionic gonadotropin (hCG) and two Fv fragments specific for the alpha and beta-subunits.
J.Mol.Biol., 289, 1999
|
|
7VQM
 
 | | GH2 beta-galacturonate AqGalA in complex with galacturonide | | Descriptor: | CHLORIDE ION, GH2 beta-galacturonate AqGalA, beta-D-galactopyranuronic acid | | Authors: | Yang, J. | | Deposit date: | 2021-10-20 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Biochemical Basis of a Marine Bacterial Glycoside Hydrolase Family 2 beta-Glycosidase with Broad Substrate Specificity
Appl.Environ.Microbiol., 88, 2022
|
|
7U9X
 
 | |
7U9Q
 
 | | Structure of PKA phosphorylated human RyR2 in the closed state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ... | | Authors: | Miotto, M.C, Marks, A.R. | | Deposit date: | 2022-03-11 | | Release date: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural analyses of human ryanodine receptor type 2 channels reveal the mechanisms for sudden cardiac death and treatment.
Sci Adv, 8, 2022
|
|
7U9T
 
 | |
7UA9
 
 | | Structure of dephosphorylated human RyR2 in the open state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, ... | | Authors: | Miotto, M.C, Marks, A.R. | | Deposit date: | 2022-03-11 | | Release date: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structural analyses of human ryanodine receptor type 2 channels reveal the mechanisms for sudden cardiac death and treatment.
Sci Adv, 8, 2022
|
|
7UA3
 
 | |
7UA1
 
 | | Structure of PKA phosphorylated human RyR2-R2474S in the closed state in the presence of ARM210 | | Descriptor: | 4-[(7-methoxy-2,3-dihydro-1,4-benzothiazepin-4(5H)-yl)methyl]benzoic acid, ADENOSINE-5'-TRIPHOSPHATE, Peptidyl-prolyl cis-trans isomerase FKBP1B, ... | | Authors: | Miotto, M.C, Marks, A.R. | | Deposit date: | 2022-03-11 | | Release date: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structural analyses of human ryanodine receptor type 2 channels reveal the mechanisms for sudden cardiac death and treatment.
Sci Adv, 8, 2022
|
|
7UA4
 
 | |
7UA5
 
 | | Structure of dephosphorylated human RyR2 in the closed state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ... | | Authors: | Miotto, M.C, Marks, A.R. | | Deposit date: | 2022-03-11 | | Release date: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural analyses of human ryanodine receptor type 2 channels reveal the mechanisms for sudden cardiac death and treatment.
Sci Adv, 8, 2022
|
|
7U9R
 
 | | Structure of PKA phosphorylated human RyR2 in the open state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, ... | | Authors: | Miotto, M.C, Marks, A.R. | | Deposit date: | 2022-03-11 | | Release date: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Structural analyses of human ryanodine receptor type 2 channels reveal the mechanisms for sudden cardiac death and treatment.
Sci Adv, 8, 2022
|
|
7U9Z
 
 | |
7VCK
 
 | |
7VJS
 
 | | Human AlkB homolog ALKBH6 in complex with Tris and Ni | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-ketoglutarate-dependent dioxygenase alkB homolog 6, NICKEL (II) ION | | Authors: | Ma, L, Chen, Z. | | Deposit date: | 2021-09-28 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Structural insights into the interactions and epigenetic functions of human nucleic acid repair protein ALKBH6.
J.Biol.Chem., 298, 2022
|
|
7VJV
 
 | | Human AlkB homolog ALKBH6 in complex with alpha-katoglutarate and Mn | | Descriptor: | 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent dioxygenase alkB homolog 6, MANGANESE (II) ION | | Authors: | Ma, L, Chen, Z. | | Deposit date: | 2021-09-29 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights into the interactions and epigenetic functions of human nucleic acid repair protein ALKBH6.
J.Biol.Chem., 298, 2022
|
|
6L3F
 
 | |
6L8K
 
 | | Structure of URT1 in complex with UTP | | Descriptor: | URIDINE 5'-TRIPHOSPHATE, UTP:RNA uridylyltransferase 1 | | Authors: | Lingru, Z. | | Deposit date: | 2019-11-06 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.999 Å) | | Cite: | Crystal structure of Arabidopsis terminal uridylyl transferase URT1.
Biochem.Biophys.Res.Commun., 524, 2020
|
|
6M07
 
 | | Crystal structure of Lp-PLA2 in complex with a novel covalent inhibitor | | Descriptor: | (2S)-2-[(E)-3-[2-(diethylamino)ethyl-[[4-[4-(trifluoromethyl)-2-[2,2,2-tris(fluoranyl)ethoxy]phenyl]phenyl]methyl]amino]-1-oxidanyl-3-oxidanylidene-prop-1-enyl]pyrrolidine-1-carboxylic acid, Platelet-activating factor acetylhydrolase | | Authors: | Hu, H.C, Xu, Y.C. | | Deposit date: | 2020-02-20 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Identification of Highly Selective Lipoprotein-Associated Phospholipase A2 (Lp-PLA2) Inhibitors by a Covalent Fragment-Based Approach.
J.Med.Chem., 63, 2020
|
|
6M08
 
 | | Crystal structure of Lp-PLA2 in complex with a novel covalent inhibitor. | | Descriptor: | (2S)-2-[(Z)-3-[2-(diethylamino)ethyl-[[4-[4-(trifluoromethyl)phenyl]phenyl]methyl]amino]-1-oxidanyl-3-oxidanylidene-prop-1-enyl]pyrrolidine-1-carboxylic acid, Platelet-activating factor acetylhydrolase | | Authors: | Hu, H.C, Xu, Y.C. | | Deposit date: | 2020-02-20 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Identification of Highly Selective Lipoprotein-Associated Phospholipase A2 (Lp-PLA2) Inhibitors by a Covalent Fragment-Based Approach.
J.Med.Chem., 63, 2020
|
|
7V1L
 
 | |
