9DE9
 
 | | Crystal Structure of HE-B11 | | Descriptor: | HE-B11 | | Authors: | Bera, A.K, Sims, J, Baker, D. | | Deposit date: | 2024-08-28 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Massively parallel assessment of designed protein solution properties using mass spectrometry and peptide barcoding.
Biorxiv, 2025
|
|
9DEA
 
 | | Crystal Structure of C3-threaded | | Descriptor: | C3_threaded | | Authors: | Bera, A.K, Sims, J, Baker, D. | | Deposit date: | 2024-08-28 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Massively parallel assessment of designed protein solution properties using mass spectrometry and peptide barcoding.
Biorxiv, 2025
|
|
9DEB
 
 | |
5ZVT
 
 | | Structure of RNA polymerase complex and genome within a dsRNA virus provides insights into the mechanisms of transcription and assembly | | Descriptor: | C-terminus of outer capsid protein VP5, Core protein VP6, MYRISTIC ACID, ... | | Authors: | Liu, H, Fang, Q, Cheng, L. | | Deposit date: | 2018-05-12 | | Release date: | 2018-07-04 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of RNA polymerase complex and genome within a dsRNA virus provides insights into the mechanisms of transcription and assembly.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4GEE
 
 | | Pyrrolopyrimidine inhibitors of DNA gyrase B and topoisomerase IV, part I: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity. | | Descriptor: | (1R,5S,6s)-3-[5-chloro-6-ethyl-2-(pyrimidin-5-yloxy)-7H-pyrrolo[2,3-d]pyrimidin-4-yl]-3-azabicyclo[3.1.0]hexan-6-amine, DNA gyrase subunit B, GLYCEROL | | Authors: | Bensen, D.C, Chen, Z, Tari, L.W. | | Deposit date: | 2012-08-01 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Pyrrolopyrimidine inhibitors of DNA gyrase B (GyrB) and topoisomerase IV (ParE). Part I: Structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
9EEC
 
 | |
7BQY
 
 | | THE CRYSTAL STRUCTURE OF COVID-19 MAIN PROTEASE IN COMPLEX WITH AN INHIBITOR N3 at 1.7 angstrom | | Descriptor: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Liu, X, Zhang, B, Jin, Z, Yang, H, Rao, Z. | | Deposit date: | 2020-03-26 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Mprofrom SARS-CoV-2 and discovery of its inhibitors.
Nature, 582, 2020
|
|
5ZKQ
 
 | | Crystal structure of the human platelet-activating factor receptor in complex with ABT-491 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-ethynyl-3-{3-fluoro-4-[(2-methyl-1H-imidazo[4,5-c]pyridin-1-yl)methyl]benzene-1-carbonyl}-N,N-dimethyl-1H-indole-1-carboxamide, Platelet-activating factor receptor,Endolysin,Endolysin,Platelet-activating factor receptor, ... | | Authors: | Cao, C, Zhao, Q, Zhang, X.C, Wu, B. | | Deposit date: | 2018-03-25 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for signal recognition and transduction by platelet-activating-factor receptor.
Nat. Struct. Mol. Biol., 25, 2018
|
|
3MB5
 
 | |
3LHD
 
 | | Crystal structure of P. abyssi tRNA m1A58 methyltransferase in complex with S-adenosyl-L-homocysteine | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SAM-dependent methyltransferase, putative | | Authors: | Guelorget, A, Golinelli-Pimpaneau, B, Wouters, J, Barbey, C. | | Deposit date: | 2010-01-22 | | Release date: | 2010-05-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Insights into the hyperthermostability and unusual region-specificity of archaeal Pyrococcus abyssi tRNA m1A57/58 methyltransferase.
Nucleic Acids Res., 38, 2010
|
|
8GZ4
 
 | | Crystal structure of MPXV phosphatase | | Descriptor: | Dual specificity protein phosphatase H1, PHOSPHATE ION | | Authors: | Yang, H.T, Wang, W, Huang, H.J, Ji, X.Y. | | Deposit date: | 2022-09-25 | | Release date: | 2023-05-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Crystal structure of monkeypox H1 phosphatase, an antiviral drug target.
Protein Cell, 14, 2023
|
|
3N6N
 
 | | crystal structure of EV71 RdRp in complex with Br-UTP | | Descriptor: | 5-bromouridine 5'-(tetrahydrogen triphosphate), NICKEL (II) ION, RNA-dependent RNA polymerase | | Authors: | Wu, Y, Lou, Z.Y, Miao, Y, Yu, Y, Rao, Z.H. | | Deposit date: | 2010-05-26 | | Release date: | 2011-06-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of EV71 RNA-dependent RNA polymerase in complex with substrate and analogue provide a drug target against the hand-foot-and-mouth disease pandemic in China.
Protein Cell, 1, 2010
|
|
5ZBZ
 
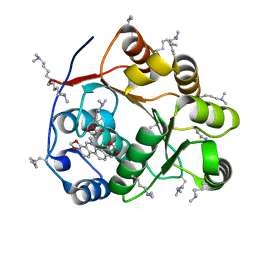 | | Crystal structure of the DEAD domain of Human eIF4A with sanguinarine | | Descriptor: | 13-methyl[1,3]benzodioxolo[5,6-c][1,3]dioxolo[4,5-i]phenanthridin-13-ium, Eukaryotic initiation factor 4A-I, MALONATE ION | | Authors: | Ding, Y, Ding, L. | | Deposit date: | 2018-02-14 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.30860257 Å) | | Cite: | Targeting the N Terminus of eIF4AI for Inhibition of Its Catalytic Recycling.
Cell Chem Biol, 26, 2019
|
|
8IS5
 
 | |
5ZKP
 
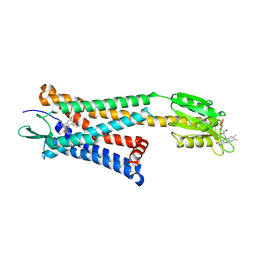 | | Crystal structure of the human platelet-activating factor receptor in complex with SR 27417 | | Descriptor: | FLAVIN MONONUCLEOTIDE, N1,N1-dimethyl-N2-[(pyridin-3-yl)methyl]-N2-{4-[2,4,6-tri(propan-2-yl)phenyl]-1,3-thiazol-2-yl}ethane-1,2-diamine, Platelet-activating factor receptor,Flavodoxin,Platelet-activating factor receptor | | Authors: | Cao, C, Zhao, Q, Zhang, X.C, Wu, B. | | Deposit date: | 2018-03-25 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural basis for signal recognition and transduction by platelet-activating-factor receptor.
Nat. Struct. Mol. Biol., 25, 2018
|
|
8HCM
 
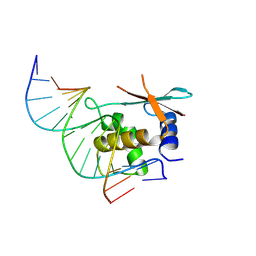 | | zebrafish IRF-11 DBD complex with DNA | | Descriptor: | DNA (5'-D(P*GP*CP*TP*TP*TP*CP*AP*CP*TP*TP*TP*CP*TP*A)-3'), DNA (5'-D(P*TP*AP*GP*AP*AP*AP*GP*TP*GP*AP*AP*AP*GP*C)-3'), Interferon regulatory factor | | Authors: | Wang, Z.X, Zhang, Y.A, Ouyang, S.Y. | | Deposit date: | 2022-11-02 | | Release date: | 2024-05-15 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal Structures of DNA-bound Fish IRF10 and IRF11 Reveal the Determinants of IFN Regulation.
J Immunol., 213, 2024
|
|
8HCL
 
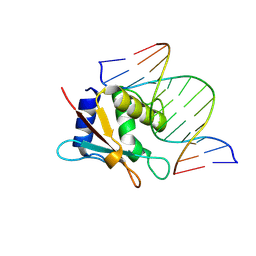 | | zebrafish IRF-10 DBD complex with DNA | | Descriptor: | DNA (5'-D(P*AP*CP*TP*TP*TP*CP*AP*CP*TP*TP*CP*A)-3'), DNA (5'-D(P*TP*GP*AP*AP*GP*TP*GP*AP*AP*AP*GP*T)-3'), Interferon regulatory factor 10 | | Authors: | Wang, Z.X, Zhang, Y.A, Ouyang, S.Y. | | Deposit date: | 2022-11-01 | | Release date: | 2024-05-15 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structures of DNA-bound Fish IRF10 and IRF11 Reveal the Determinants of IFN Regulation.
J Immunol., 213, 2024
|
|
8HCS
 
 | | zebrafish IRF-11 DBD | | Descriptor: | Interferon regulatory factor | | Authors: | Wang, Z.X, Zhang, Y.A, Ouyang, S.Y. | | Deposit date: | 2022-11-03 | | Release date: | 2024-05-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structures of DNA-bound Fish IRF10 and IRF11 Reveal the Determinants of IFN Regulation.
J Immunol., 213, 2024
|
|
4EYC
 
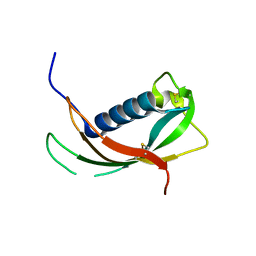 | | Crystal structure of the cathelin-like domain of human cathelicidin LL-37 (hCLD) | | Descriptor: | Cathelicidin antimicrobial peptide | | Authors: | Pazgier, M, Pozharski, E, Toth, E, Lu, W. | | Deposit date: | 2012-05-01 | | Release date: | 2013-02-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Analysis of the Pro-Domain of Human Cathelicidin, LL-37.
Biochemistry, 52, 2013
|
|
8HNW
 
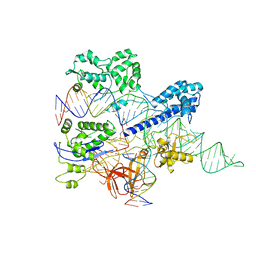 | | Crystal structure of HpaCas9-sgRNA surveillance complex bound to double-stranded DNA | | Descriptor: | CRISPR-associated endonuclease Cas9, Non-target strand, Target strand, ... | | Authors: | Sun, W, Cheng, Z, Wang, Y. | | Deposit date: | 2022-12-08 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | AcrIIC4 inhibits type II-C Cas9 by preventing R-loop formation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8HNS
 
 | | Crystal structure of an anti-CRISPR protein AcrIIC4 in apo form | | Descriptor: | GLYCEROL, anti-CRISPR protein AcrIIC4 | | Authors: | Sun, W, Cheng, Z, Yang, J, Wang, Y. | | Deposit date: | 2022-12-08 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | AcrIIC4 inhibits type II-C Cas9 by preventing R-loop formation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8HNT
 
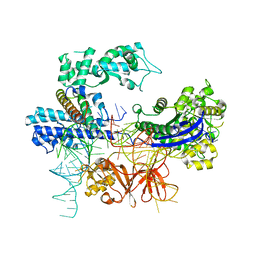 | |
7UBF
 
 | |
7UBD
 
 | |
7UBI
 
 | |
