5K8H
 
 | |
7MK2
 
 | | CryoEM Structure of NPR1 | | Descriptor: | Regulatory protein NPR1, ZINC ION | | Authors: | Kumar, S, Zhou, Y, Dillard, L, Borgnia, M, Bartesaghi, A, Zhou, P. | | Deposit date: | 2021-04-21 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
6V97
 
 | | Kindlin-3 double deletion mutant short form | | Descriptor: | Fermitin family homolog 3 | | Authors: | Xu, Z, Zhang, T.L, Xu, Z, Sun, J.J, Ding, J.P, Ma, Y.Q. | | Deposit date: | 2019-12-13 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.381 Å) | | Cite: | Structure basis of the FERM domain of kindlin-3 in supporting integrin alpha IIb beta 3 activation in platelets.
Blood Adv, 4, 2020
|
|
6V9G
 
 | | Kindlin-3 double deletion mutant long form | | Descriptor: | Fermitin family homolog 3 | | Authors: | Xu, Z, Zhang, T.L, Xu, Z, Sun, J.J, Ding, J.P, Ma, Y.Q. | | Deposit date: | 2019-12-13 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure basis of the FERM domain of kindlin-3 in supporting integrin alpha IIb beta 3 activation in platelets.
Blood Adv, 4, 2020
|
|
3K9X
 
 | | X-ray crystal structure of human fxa in complex with (S)-N-((2-METHYLBENZOFURAN-5-YLAMINO)(2-OXO-1-(2-OXO-2- (PYRROLIDIN-1-YL)ETHYL)AZEPAN-3- YLAMINO)METHYLENE)NICOTINAMIDE | | Descriptor: | CALCIUM ION, GLYCEROL, N-{N'-(2-methyl-1-benzofuran-5-yl)-N-[(3S)-2-oxo-1-(2-oxo-2-pyrrolidin-1-ylethyl)azepan-3-yl]carbamimidoyl}pyridine-3-carboxamide, ... | | Authors: | Klei, H.E, Kish, K, Ghosh, K, Rushith, A. | | Deposit date: | 2009-10-16 | | Release date: | 2009-12-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Aroylguanidine-based factor Xa inhibitors: the discovery of BMS-344577
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3HPT
 
 | | Crystal structure of human FxA in complex with (S)-2-cyano-1-(2-methylbenzofuran-5-yl)-3-(2-oxo-1-(2-oxo-2-(pyrrolidin-1-yl)ethyl)azepan-3-yl)guanidine | | Descriptor: | 1-cyano-2-(2-methyl-1-benzofuran-5-yl)-3-[(3S)-2-oxo-1-(2-oxo-2-pyrrolidin-1-ylethyl)azepan-3-yl]guanidine, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Klei, H.E, Ghosh, K, Rushith, A, Kish, K. | | Deposit date: | 2009-06-04 | | Release date: | 2009-11-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Cyanoguanidine-based lactam derivatives as a novel class of orally bioavailable factor Xa inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
5UL5
 
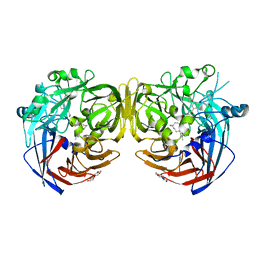 | | Crystal structure of RPE65 in complex with MB-004 and palmitate | | Descriptor: | (1R)-3-amino-1-{3-[(2-propylpentyl)oxy]phenyl}propan-1-ol, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, FE (II) ION, ... | | Authors: | Kiser, P.D, Palczewski, K. | | Deposit date: | 2017-01-24 | | Release date: | 2017-05-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational Tuning of Visual Cycle Modulator Pharmacodynamics.
J. Pharmacol. Exp. Ther., 362, 2017
|
|
5ULG
 
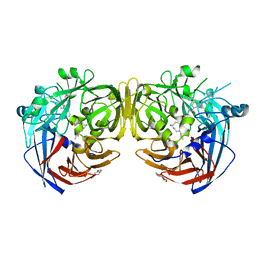 | |
3GYN
 
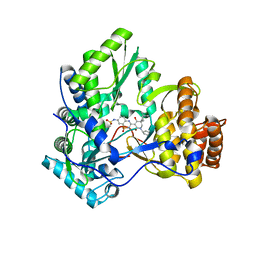 | | Crystal structure of HCV NS5B polymerase with a novel monocyclic dihydropyridinone inhibitor | | Descriptor: | N-{3-[(5R)-1-cyclopentyl-4-hydroxy-5-methyl-5-(3-methylbutyl)-2-oxo-1,2,5,6-tetrahydropyridin-3-yl]-1,1-dioxido-4H-1,2,4-benzothiadiazin-7-yl}methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Zhao, Q, Showalter, R.E, Han, Q, Kissinger, C.R. | | Deposit date: | 2009-04-04 | | Release date: | 2009-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 5,5'- and 6,6'-dialkyl-5,6-dihydro-1H-pyridin-2-ones as potent inhibitors of HCV NS5B polymerase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
6JNL
 
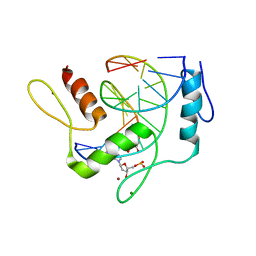 | | REF6 ZnF2-4-NAC004 complex | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, DNA (5'-D(*CP*AP*AP*AP*AP*CP*AP*GP*AP*GP*A)-3'), DNA (5'-D(*TP*TP*CP*TP*CP*TP*GP*TP*TP*TP*TP*G)-3'), ... | | Authors: | Yao, Q.Q, Wu, B.X, Ma, J.B. | | Deposit date: | 2019-03-17 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | DNA methylation repels targeting of Arabidopsis REF6.
Nat Commun, 10, 2019
|
|
3DBN
 
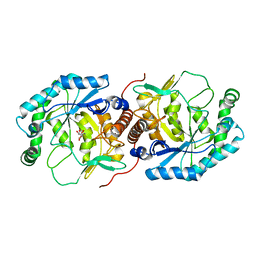 | | Crystal structure of the Streptoccocus suis serotype2 D-mannonate dehydratase in complex with its substrate | | Descriptor: | D-MANNONIC ACID, MANGANESE (II) ION, Mannonate dehydratase | | Authors: | Peng, H, Zhang, Q, Gao, F, Gao, G.F. | | Deposit date: | 2008-06-02 | | Release date: | 2009-06-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of Streptococcus suis mannonate dehydratase (ManD) and its complex with substrate: genetic and biochemical evidence for a catalytic mechanism
J.Bacteriol., 191, 2009
|
|
6JNM
 
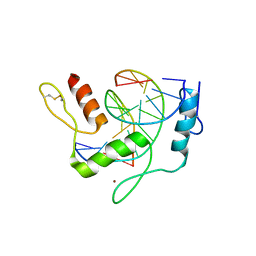 | | REF6 ZnF2-4-NAC004-mC3 complex | | Descriptor: | DNA (5'-D(*CP*AP*AP*AP*AP*CP*AP*GP*AP*GP*AP*A)-3'), DNA (5'-D(*TP*TP*CP*TP*(5CM)P*TP*GP*TP*TP*TP*TP*G)-3'), Lysine-specific demethylase REF6, ... | | Authors: | Yao, Q.Q, Wu, B.X, Ma, J.B. | | Deposit date: | 2019-03-17 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | DNA methylation repels targeting of Arabidopsis REF6.
Nat Commun, 10, 2019
|
|
6JNN
 
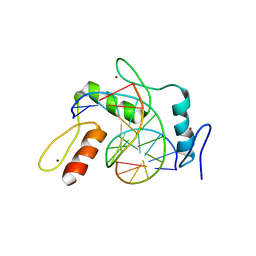 | | REF6 ZnF2-4-NAC004-mC1 complex | | Descriptor: | DNA (5'-D(*CP*AP*AP*AP*AP*CP*AP*GP*AP*GP*AP*A)-3'), DNA (5'-D(*TP*TP*(5CM)P*TP*CP*TP*GP*TP*TP*TP*TP*G)-3'), Lysine-specific demethylase REF6, ... | | Authors: | Yao, Q.Q, Wu, B.X, Ma, J.B. | | Deposit date: | 2019-03-17 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | DNA methylation repels targeting of Arabidopsis REF6.
Nat Commun, 10, 2019
|
|
5F59
 
 | | The crystal structure of MLL3 SET domain | | Descriptor: | Histone-lysine N-methyltransferase 2C, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Li, Y, Lei, M, Chen, Y. | | Deposit date: | 2015-12-04 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural basis for activity regulation of MLL family methyltransferases.
Nature, 530, 2016
|
|
6NYH
 
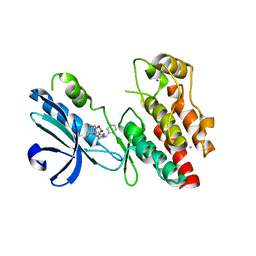 | | Structure of human RIPK1 kinase domain in complex with GNE684 | | Descriptor: | (5S)-N-[(3S)-7-methoxy-1-methyl-2-oxo-2,3,4,5-tetrahydro-1H-pyrido[3,4-b]azepin-3-yl]-5-phenyl-6,7-dihydro-5H-pyrrolo[1,2-b][1,2,4]triazole-2-carboxamide, IODIDE ION, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Fong, R, Lupardus, P.J. | | Deposit date: | 2019-02-11 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | RIP1 inhibition blocks inflammatory diseases but not tumor growth or metastases.
Cell Death Differ., 27, 2020
|
|
5F6L
 
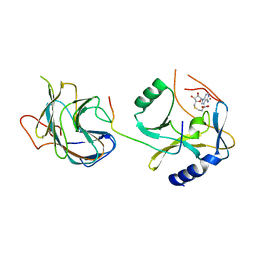 | | The crystal structure of MLL1 (N3861I/Q3867L) in complex with RbBP5 and Ash2L | | Descriptor: | Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Li, Y, Lei, M, Chen, Y. | | Deposit date: | 2015-12-06 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for activity regulation of MLL family methyltransferases.
Nature, 530, 2016
|
|
5F5E
 
 | |
5F6K
 
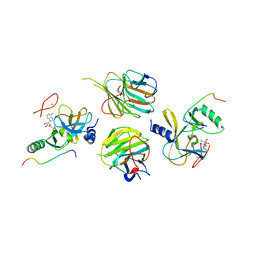 | | Crystal structure of the MLL3-Ash2L-RbBP5 complex | | Descriptor: | Histone-lysine N-methyltransferase 2C, Retinoblastoma-binding protein 5, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Li, Y, Lei, M, Chen, Y. | | Deposit date: | 2015-12-06 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.411 Å) | | Cite: | Structural basis for activity regulation of MLL family methyltransferases.
Nature, 530, 2016
|
|
6UD7
 
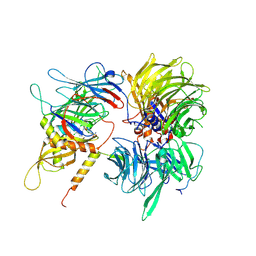 | | Crystal structure of full-length human DCAF15-DDB1(deltaBPB)-DDA1-RBM39 in complex with indisulam | | Descriptor: | DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, DNA damage-binding protein 1,DNA damage-binding protein 1, ... | | Authors: | Bussiere, D.E, Shu, W, Xie, L, Knapp, M. | | Deposit date: | 2019-09-18 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of indisulam-mediated RBM39 recruitment to DCAF15 E3 ligase complex.
Nat.Chem.Biol., 16, 2020
|
|
8TQD
 
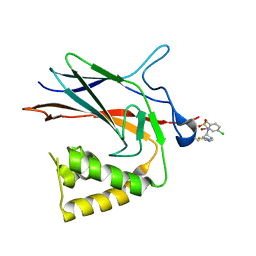 | | NF-Kappa-B1 Bound with a Covalent Inhibitor | | Descriptor: | 1-(2-bromo-4-chlorophenyl)-N-{(3S)-1-[(E)-iminomethyl]pyrrolidin-3-yl}methanesulfonamide, Nuclear factor NF-kappa-B p105 subunit | | Authors: | Hilbert, B.J. | | Deposit date: | 2023-08-07 | | Release date: | 2024-04-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | DrugMap: A quantitative pan-cancer analysis of cysteine ligandability.
Cell, 187, 2024
|
|
6UE5
 
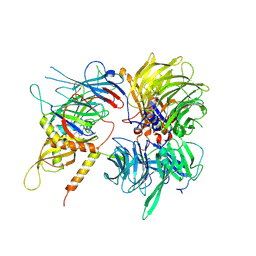 | | Crystal structure of full-length human DCAF15-DDB1-deltaPBP-DDA1-RBM39 in complex with 4-(aminomethyl)-N-(3-cyano-4-methyl-1H-indol-7-yl)benzenesulfonamide | | Descriptor: | 4-(aminomethyl)-N-(3-cyano-4-methyl-1H-indol-7-yl)benzene-1-sulfonamide, DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, ... | | Authors: | Knapp, M.S, Shu, W, Xie, L, Bussiere, D.E. | | Deposit date: | 2019-09-20 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural basis of indisulam-mediated RBM39 recruitment to DCAF15 E3 ligase complex.
Nat.Chem.Biol., 16, 2020
|
|
6LPF
 
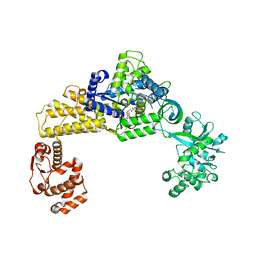 | | The crystal structure of human cytoplasmic LRS | | Descriptor: | 2'-(L-NORVALYL)AMINO-2'-DEOXYADENOSINE, 5'-O-(L-leucylsulfamoyl)adenosine, GLYCEROL, ... | | Authors: | Liu, R.J, Long, T, Li, H, Li, J, Zhao, J.H, Lin, J.Z, Palencia, A, Wang, M.Z, Cusack, S, Wang, E.D. | | Deposit date: | 2020-01-10 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Molecular basis of the multifaceted functions of human leucyl-tRNA synthetase in protein synthesis and beyond.
Nucleic Acids Res., 48, 2020
|
|
6PBT
 
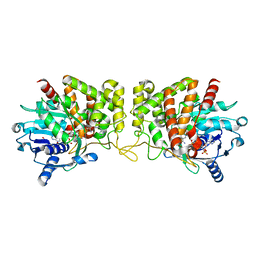 | | Pseudopaline Dehydrogenase with (R)-Pseudopaline Soaked 2 hours | | Descriptor: | 1,2-ETHANEDIOL, N-[(3S)-3-amino-3-carboxypropyl]-L-histidine, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | McFarlane, J.S, Lamb, A.L. | | Deposit date: | 2019-06-14 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Staphylopine and pseudopaline dehydrogenase from bacterial pathogens catalyze reversible reactions and produce stereospecific metallophores.
J.Biol.Chem., 294, 2019
|
|
6LR6
 
 | | The crystal structure of human cytoplasmic LRS | | Descriptor: | 4-Chloro-3-aminomethyl-7-[ethoxy]-3H-benzo[C][1,2]oxaborol-1-ol modified adenosine, 5'-O-(L-leucylsulfamoyl)adenosine, Leucine--tRNA ligase, ... | | Authors: | Liu, R.J, Long, T, Li, H, Li, J, Zhao, J.H, Lin, J.Z, Palencia, A, Wang, M.Z, Cusack, S, Wang, E.D. | | Deposit date: | 2020-01-15 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.009 Å) | | Cite: | Molecular basis of the multifaceted functions of human leucyl-tRNA synthetase in protein synthesis and beyond.
Nucleic Acids Res., 48, 2020
|
|
6PBN
 
 | | Pseudopaline Dehydrogenase with (R)-Pseudopaline Soaked 1 hour | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, N-[(3S)-3-amino-3-carboxypropyl]-L-histidine, ... | | Authors: | McFarlane, J.S, Lamb, A.L. | | Deposit date: | 2019-06-14 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Staphylopine and pseudopaline dehydrogenase from bacterial pathogens catalyze reversible reactions and produce stereospecific metallophores.
J.Biol.Chem., 294, 2019
|
|
