1KA4
 
 | | Structure of Pyrococcus furiosus carboxypeptidase Nat-Pb | | Descriptor: | LEAD (II) ION, M32 carboxypeptidase | | Authors: | Arndt, J.W, Hao, B, Ramakrishnan, V, Cheng, T, Chan, S.I, Chan, M.K. | | Deposit date: | 2001-10-31 | | Release date: | 2002-11-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of a Novel Carboxypeptidase from the Hyperthermophilic Archaeon Pyrococcus furiosus
Structure, 10, 2002
|
|
1DP9
 
 | | CRYSTAL STRUCTURE OF IMIDAZOLE-BOUND FIXL HEME DOMAIN | | Descriptor: | FIXL PROTEIN, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gong, W, Hao, B, Chan, M.K. | | Deposit date: | 1999-12-24 | | Release date: | 2000-12-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | New mechanistic insights from structural studies of the oxygen-sensing domain of Bradyrhizobium japonicum FixL.
Biochemistry, 39, 2000
|
|
6O59
 
 | |
1DRM
 
 | | CRYSTAL STRUCTURE OF THE LIGAND FREE BJFIXL HEME DOMAIN | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, SENSOR PROTEIN FIXL | | Authors: | Gong, W, Hao, B, Mansy, S.S, Gonzalez, G, Gilles-Gonzalez, M.A, Chan, M.K. | | Deposit date: | 2000-01-06 | | Release date: | 2000-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a biological oxygen sensor: a new mechanism for heme-driven signal transduction.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1DFF
 
 | | PEPTIDE DEFORMYLASE | | Descriptor: | PEPTIDE DEFORMYLASE, ZINC ION | | Authors: | Chan, M.K, Gong, W, Rajagopalan, P.T.R, Hao, B, Tsai, C.M, Pei, D. | | Deposit date: | 1997-08-19 | | Release date: | 1998-09-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Crystal structure of the Escherichia coli peptide deformylase.
Biochemistry, 36, 1997
|
|
3CF4
 
 | | Structure of the CODH component of the M. barkeri ACDS complex | | Descriptor: | ACETIC ACID, Acetyl-CoA decarboxylase/synthase alpha subunit, Acetyl-CoA decarboxylase/synthase epsilon subunit, ... | | Authors: | Gong, W, Hao, B, Wei, Z, Ferguson Jr, D.J, Tallant, T, Krzycki, J.A, Chan, M.K. | | Deposit date: | 2008-03-01 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the alpha2 epsilon2 Ni-dependent CO dehydrogenase component of the Methanosarcina barkeri acetyl-CoA decarboxylase/synthase complex
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
1XEO
 
 | | High Resolution Crystals Structure of Cobalt- Peptide Deformylase Bound To Formate | | Descriptor: | COBALT (II) ION, FORMIC ACID, Peptide deformylase | | Authors: | Jain, R, Hao, B, Liu, R.-P, Chan, M.K. | | Deposit date: | 2004-09-10 | | Release date: | 2005-03-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of E. coli peptide deformylase bound to formate: insight into the preference for Fe2+ over Zn2+ as the active site metal
J.Am.Chem.Soc., 127, 2005
|
|
1XEM
 
 | | High Resolution Crystal Structure of Escherichia coli Zinc- Peptide Deformylase bound to formate | | Descriptor: | FORMIC ACID, Peptide deformylase, ZINC ION | | Authors: | Jain, R, Hao, B, Liu, R.-P, Chan, M.K. | | Deposit date: | 2004-09-10 | | Release date: | 2005-03-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structures of E. coli peptide deformylase bound to formate: insight into the preference for Fe2+ over Zn2+ as the active site metal
J.Am.Chem.Soc., 127, 2005
|
|
1XEN
 
 | | High Resolution Crystal Structure of Escherichia coli Iron- Peptide Deformylase Bound To Formate | | Descriptor: | FE (III) ION, FORMIC ACID, Peptide deformylase | | Authors: | Jain, R, Hao, B, Liu, R.-P, Chan, M.K. | | Deposit date: | 2004-09-10 | | Release date: | 2005-03-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of E. coli peptide deformylase bound to formate: insight into the preference for Fe2+ over Zn2+ as the active site metal
J.Am.Chem.Soc., 127, 2005
|
|
3N54
 
 | | Crystal Structure of the GerBC protein | | Descriptor: | CHLORIDE ION, SULFATE ION, Spore germination protein B3 | | Authors: | Li, Y, Setlow, B, Setlow, P, Hao, B. | | Deposit date: | 2010-05-24 | | Release date: | 2010-08-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the GerBC Component of a Bacillus subtilis Spore Germinant Receptor.
J.Mol.Biol., 402, 2010
|
|
7UR2
 
 | |
4O8W
 
 | | Crystal Structure of the GerD spore germination protein | | Descriptor: | Spore germination protein | | Authors: | Li, Y, Jin, K, Ghosh, S, Devarakonda, P, Carlson, K, Davis, A, Stewart, K, Cammett, E, Rossi, P.P, Setlow, B, Lu, M, Setlow, P, Hao, B. | | Deposit date: | 2013-12-30 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Structural and Functional Analysis of the GerD Spore Germination Protein of Bacillus Species.
J.Mol.Biol., 426, 2014
|
|
3L2O
 
 | |
8F2E
 
 | |
5VZU
 
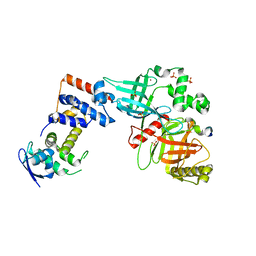 | | Crystal structure of the Skp1-FBXO31-cyclin D1 complex | | Descriptor: | Cyclin D1, F-box only protein 31, PHOSPHATE ION, ... | | Authors: | Li, Y, Jin, K, Hao, B. | | Deposit date: | 2017-05-29 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of the phosphorylation-independent recognition of cyclin D1 by the SCFFBXO31 ubiquitin ligase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5V4B
 
 | | Crystal structure of the Skp1-FBXW7-DISC1 complex | | Descriptor: | DISC1 peptide, F-box/WD repeat-containing protein 7, IMIDAZOLE, ... | | Authors: | Li, Y, Baillie, G.S, Hao, B. | | Deposit date: | 2017-03-08 | | Release date: | 2017-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | FBXW7 regulates DISC1 stability via the ubiquitin-proteosome system.
Mol. Psychiatry, 23, 2018
|
|
5VZT
 
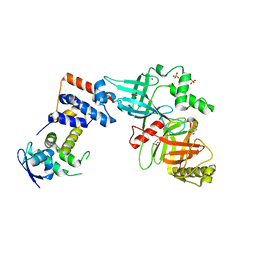 | | Crystal structure of the Skp1-FBXO31 complex | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, F-box only protein 31, PHOSPHATE ION, ... | | Authors: | Li, Y, Jin, K, Hao, B. | | Deposit date: | 2017-05-29 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of the phosphorylation-independent recognition of cyclin D1 by the SCFFBXO31 ubiquitin ligase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2M19
 
 | | Solution structure of the Haloferax volcanii HVO 2177 protein | | Descriptor: | Molybdopterin converting factor subunit 1 | | Authors: | Li, Y, Maciejewski, M.W, Martin, J, Jin, K, Zhang, Y, Lu, M, Maupin-Furlow, J.A, Hao, B. | | Deposit date: | 2012-11-21 | | Release date: | 2013-08-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Crystal structure of the ubiquitin-like small archaeal modifier protein 2 from Haloferax volcanii.
Protein Sci., 22, 2013
|
|
6BCD
 
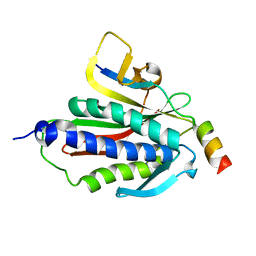 | | Crystal structure of Rev7-K44A/R124A/A135D in complex with Rev3-RBM2 (residues 1988-2014) | | Descriptor: | DNA polymerase zeta catalytic subunit, Mitotic spindle assembly checkpoint protein MAD2B | | Authors: | Rizzo, A.A, Hao, B, Li, Y, Korzhnev, D.M. | | Deposit date: | 2017-10-20 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Rev7 dimerization is important for assembly and function of the Rev1/Pol zeta translesion synthesis complex.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BC8
 
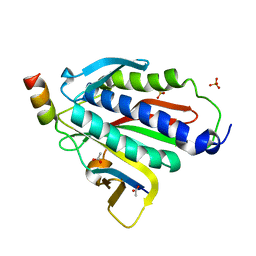 | | Crystal structure of Rev7-R124A/Rev3-RBM2 (residues 1988-2014) complex | | Descriptor: | ACETATE ION, DNA polymerase zeta catalytic subunit, Mitotic spindle assembly checkpoint protein MAD2B, ... | | Authors: | Rizzo, A.A, Hao, B, Li, Y, Korzhnev, D.M. | | Deposit date: | 2017-10-20 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Rev7 dimerization is important for assembly and function of the Rev1/Pol zeta translesion synthesis complex.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BI7
 
 | | Crystal structure of Rev7-WT/Rev3 as a monomer under high-salt conditions | | Descriptor: | DNA polymerase zeta catalytic subunit, Mitotic spindle assembly checkpoint protein MAD2B | | Authors: | Rizzo, A.A, Korzhnev, D.M, Hao, B, Li, Y. | | Deposit date: | 2017-11-01 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Rev7 dimerization is important for assembly and function of the Rev1/Pol zeta translesion synthesis complex.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1ZYS
 
 | | Co-crystal structure of Checkpoint Kinase Chk1 with a pyrrolo-pyridine inhibitor | | Descriptor: | N-{5-[4-(4-METHYLPIPERAZIN-1-YL)PHENYL]-1H-PYRROLO[2,3-B]PYRIDIN-3-YL}NICOTINAMIDE, SULFATE ION, Serine/threonine-protein kinase Chk1, ... | | Authors: | Stavenger, R.A, Zhao, B, Zhou, B.-B.S, Brown, M.J, Lee, D, Holt, D.A. | | Deposit date: | 2005-06-10 | | Release date: | 2006-06-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Pyrrolo[2,3-b]pyridines Inhibit the Checkpoint Kinase Chk1
To be Published
|
|
4X0W
 
 | | The crystal structure of mupain-1-17 in complex with murinised human uPA | | Descriptor: | SULFATE ION, Urokinase-type plasminogen activator, mupain-1-17, ... | | Authors: | Jiang, L, Zhao, B, Xu, P, Andreasen, P, Huang, M. | | Deposit date: | 2014-11-24 | | Release date: | 2015-10-21 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Distinctive binding modes and inhibitory mechanisms of two peptidic inhibitors of urokinase-type plasminogen activator with isomeric P1 residues.
Int.J.Biochem.Cell Biol., 62, 2015
|
|
4X1P
 
 | | The crystal structure of mupain-1-17 in complex with murinised human uPA at pH4.6 | | Descriptor: | MUPAIN-1-17, SULFATE ION, TRIETHYLENE GLYCOL, ... | | Authors: | Jiang, L, Zhao, B, Xu, P, Andreasen, P, Huang, M. | | Deposit date: | 2014-11-25 | | Release date: | 2015-10-21 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Distinctive binding modes and inhibitory mechanisms of two peptidic inhibitors of urokinase-type plasminogen activator with isomeric P1 residues.
Int.J.Biochem.Cell Biol., 62, 2015
|
|
1BGO
 
 | | CRYSTAL STRUCTURE OF CYSTEINE PROTEASE HUMAN CATHEPSIN K IN COMPLEX WITH A COVALENT PEPTIDOMIMETIC INHIBITOR | | Descriptor: | 1-[2-(3-BIPHENYL)-4-METHYLVALERYL)]AMINO-3-(2-PYRIDYLSULFONYL)AMINO-2-PROPANONE, CATHEPSIN K | | Authors: | Desjarlais, R.L, Yamashita, D.S, Oh, H.-J, Bondinell, W.E, Uzinskas, I.N, Erhard, K.F, Allen, A.C, Haltiwanger, R.C, Zhao, B, Smith, W.W, Abdel-Meguid, S.S, D'Alessio, K, Janson, C.A, Mcqueney, M.S, Tomaszek, T.A, Levy, M.A, Veber, D.F. | | Deposit date: | 1998-05-29 | | Release date: | 1999-06-08 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Use of X-Ray Co-Crystal Structures and Molecular Modeling to Design Potent and Selective Non-Peptide Inhibitors of Cathepsin K
J.Am.Chem.Soc., 120, 1998
|
|
