9J0Y
 
 | |
9J0Z
 
 | |
8I39
 
 | | Cryo-EM structure of abscisic acid transporter AtABCG25 with ABA | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ABC transporter G family member 25 | | Authors: | Huang, X, Zhang, X, Zhang, P. | | Deposit date: | 2023-01-16 | | Release date: | 2023-09-13 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Cryo-EM structure and molecular mechanism of abscisic acid transporter ABCG25.
Nat.Plants, 9, 2023
|
|
3DW8
 
 | | Structure of a Protein Phosphatase 2A Holoenzyme with B55 subunit | | Descriptor: | MANGANESE (II) ION, Serine/threonine-protein phosphatase 2A 55 kDa regulatory subunit B alpha isoform, Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform, ... | | Authors: | Xu, Y, Chen, Y, Zhang, P, Jeffrey, P.D, Shi, Y. | | Deposit date: | 2008-07-21 | | Release date: | 2008-10-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of a protein phosphatase 2A holoenzyme: insights into B55-mediated Tau dephosphorylation.
Mol.Cell, 31, 2008
|
|
3E9F
 
 | | Crystal structure short-form (residue1-113) of Eaf3 chromo domain | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Chromatin modification-related protein EAF3 | | Authors: | Sun, B, Hong, J, Zhang, P, Lin, D, Ding, J. | | Deposit date: | 2008-08-22 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis of the Interaction of Saccharomyces cerevisiae Eaf3 Chromo Domain with Methylated H3K36
J.Biol.Chem., 283, 2008
|
|
3E9G
 
 | | Crystal structure long-form (residue1-124) of Eaf3 chromo domain | | Descriptor: | Chromatin modification-related protein EAF3 | | Authors: | Sun, B, Hong, J, Zhang, P, Lin, D, Ding, J. | | Deposit date: | 2008-08-22 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Basis of the Interaction of Saccharomyces cerevisiae Eaf3 Chromo Domain with Methylated H3K36
J.Biol.Chem., 283, 2008
|
|
8XAT
 
 | |
8XAS
 
 | | Crystal structure of AtARR1-DBD in complex with a DNA fragment | | Descriptor: | DNA (50-MER), Two-component response regulator ARR1 | | Authors: | Li, J.X, Zhou, C.M, zhang, P, Wang, J.W. | | Deposit date: | 2023-12-05 | | Release date: | 2024-01-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.346 Å) | | Cite: | The structure of B-ARR reveals the molecular basis of transcriptional activation by cytokinin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8RI6
 
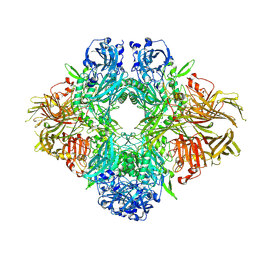 | |
8RI7
 
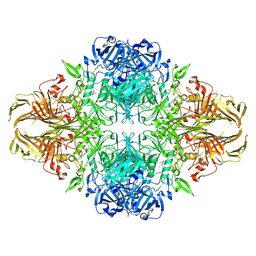 | | Beta-galactosidase (LacZ) in complex with glycosyrin from Pseudomonas syringae. | | Descriptor: | (2S,3R,4S)-2-[bis(oxidanyl)methyl]pyrrolidine-3,4-diol, Beta-galactosidase, MAGNESIUM ION, ... | | Authors: | Hardenbrook, N.J, Sheng, Y, Zhang, P. | | Deposit date: | 2023-12-18 | | Release date: | 2025-01-01 | | Last modified: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (1.93 Å) | | Cite: | Identification of a novel inhibitor for beta-galactosidase from Pseudomonas syringae.
To Be Published
|
|
8RI8
 
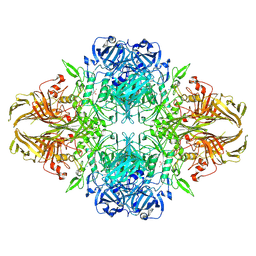 | | Beta-galactosidase (LacZ) in complex with synthetic glycosyrin. | | Descriptor: | (2S,3R,4S)-2-[bis(oxidanyl)methyl]pyrrolidine-3,4-diol, Beta-galactosidase, MAGNESIUM ION, ... | | Authors: | Hardenbrook, N.J, Sheng, Y, Zhang, P. | | Deposit date: | 2023-12-18 | | Release date: | 2025-01-01 | | Last modified: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (1.42 Å) | | Cite: | Identification of a novel inhibitor for beta-galactosidase from Pseudomonas syringae.
To Be Published
|
|
6Y9X
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-13,7) | | Descriptor: | Gag-Pol polyprotein, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2020-03-10 | | Release date: | 2020-08-19 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6Y9Z
 
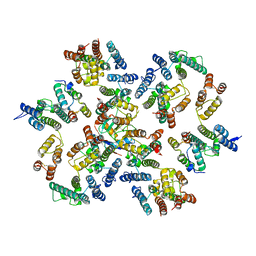 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-13,9) | | Descriptor: | Gag-Pol polyprotein, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2020-03-10 | | Release date: | 2020-08-19 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6WJF
 
 | | PKA RIIbeta holoenzyme with DnaJB1-PKAc fusion in fibrolamellar hepatoceullar carcinoma | | Descriptor: | DnaJ homolog subfamily B member 1,cAMP-dependent protein kinase catalytic subunit alpha fusion, cAMP-dependent protein kinase type II-beta regulatory subunit | | Authors: | Lu, T.-W, Aoto, P.C, Weng, J.-H, Nielsen, C, Cash, J.N, Hall, J, Zhang, P, Simon, S.M, Cianfrocco, M.A, Taylor, S.S. | | Deposit date: | 2020-04-13 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Structural analyses of the PKA RII beta holoenzyme containing the oncogenic DnaJB1-PKAc fusion protein reveal protomer asymmetry and fusion-induced allosteric perturbations in fibrolamellar hepatocellular carcinoma.
Plos Biol., 18, 2020
|
|
6WJG
 
 | | PKA RIIbeta holoenzyme with DnaJB1-PKAc fusion in fibrolamellar hepatoceullar carcinoma | | Descriptor: | DnaJ homolog subfamily B member 1, cAMP-dependent protein kinase catalytic subunit alpha fusion, cAMP-dependent protein kinase type II-beta regulatory subunit | | Authors: | Lu, T.-W, Aoto, P.C, Weng, J.-H, Nielsen, C, Cash, J.N, Hall, J, Zhang, P, Simon, S.M, Cianfrocco, M.A, Taylor, S.S. | | Deposit date: | 2020-04-13 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structural analyses of the PKA RII beta holoenzyme containing the oncogenic DnaJB1-PKAc fusion protein reveal protomer asymmetry and fusion-induced allosteric perturbations in fibrolamellar hepatocellular carcinoma.
Plos Biol., 18, 2020
|
|
6Y9V
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-8,13) | | Descriptor: | Gag-Pol polyprotein, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2020-03-10 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5UP4
 
 | |
6XF7
 
 | | SLP | | Descriptor: | Lambda 1 protein | | Authors: | Sutton, G, Sun, D.P, Fu, X.F, Kotecha, A, Hecksel, G.W, Clare, D.K, Zhang, P, Stuart, D, Boyce, M. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Assembly intermediates of orthoreovirus captured in the cell.
Nat Commun, 11, 2020
|
|
6Y9Y
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-7,13) | | Descriptor: | Gag-Pol polyprotein, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2020-03-10 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6Y9W
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-13,8) | | Descriptor: | Gag-Pol polyprotein, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2020-03-10 | | Release date: | 2020-08-19 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8IAB
 
 | | The Arabidopsis CLCa transporter bound with chloride, ATP and PIP2 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, Chloride channel protein CLC-a, ... | | Authors: | Yang, Z, Zhang, X, Zhang, P. | | Deposit date: | 2023-02-08 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Molecular mechanism underlying regulation of Arabidopsis CLCa transporter by nucleotides and phospholipids.
Nat Commun, 14, 2023
|
|
8IAD
 
 | | The Arabidopsis CLCa transporter bound with nitrate, ATP and PIP2 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chloride channel protein CLC-a, MAGNESIUM ION, ... | | Authors: | Yang, Z, Zhang, X, Zhang, P. | | Deposit date: | 2023-02-08 | | Release date: | 2023-08-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Molecular mechanism underlying regulation of Arabidopsis CLCa transporter by nucleotides and phospholipids.
Nat Commun, 14, 2023
|
|
5X41
 
 | | 3.5A resolution structure of a cobalt energy-coupling factor transporter using LCP method-CbiMQO | | Descriptor: | Cobalt ABC transporter ATP-binding protein, Cobalt transport protein CbiM, Uncharacterized protein CbiQ | | Authors: | Bao, Z, Qi, X, Zhao, W, Li, D, Zhang, P. | | Deposit date: | 2017-02-09 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | Structure and mechanism of a group-I cobalt energy coupling factor transporter
Cell Res., 27, 2017
|
|
5X40
 
 | | Structure of a CbiO dimer bound with AMPPCP | | Descriptor: | Cobalt ABC transporter ATP-binding protein, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Bao, Z, Qi, X, Wang, J, Zhang, P. | | Deposit date: | 2017-02-09 | | Release date: | 2017-04-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and mechanism of a group-I cobalt energy coupling factor transporter
Cell Res., 27, 2017
|
|
5XN6
 
 | | Heterodimer crystal structure of geranylgeranyl diphosphate synthases 1 with GGPPS Recruiting Protein(OsGRP) from Oryza sativa | | Descriptor: | Os02g0668100 protein, Os07g0580900 protein | | Authors: | Wang, C, Zhou, F, Lu, S, Zhang, P. | | Deposit date: | 2017-05-18 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.598 Å) | | Cite: | A recruiting protein of geranylgeranyl diphosphate synthase controls metabolic flux toward chlorophyll biosynthesis in rice
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
