6UZA
 
 | | Cryo-EM structure of human TRPC6 in complex with antagonist AM-1473 | | Descriptor: | 2-[[(2~{S})-2-decanoyloxypropoxy]-oxidanyl-phosphoryl]oxyethyl-trimethyl-azanium, 4-({(1R,2R)-2-[(3R)-3-aminopiperidin-1-yl]-2,3-dihydro-1H-inden-1-yl}oxy)benzonitrile, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Bai, Y, Yu, X, Huang, X, Chen, H. | | Deposit date: | 2019-11-14 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural basis for pharmacological modulation of the TRPC6 channel.
Elife, 9, 2020
|
|
2KI0
 
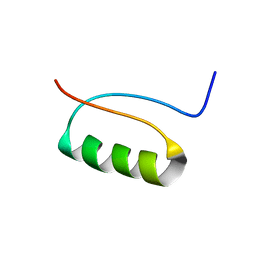 | | NMR Structure of a de novo designed beta alpha beta | | Descriptor: | DS119 | | Authors: | Liang, H, Chen, H, Fan, K, Wei, P, Guo, X, Jin, C, Zeng, C, Tang, C, Lai, L. | | Deposit date: | 2009-04-18 | | Release date: | 2009-10-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | De novo design of a beta alpha beta motif.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
1EC6
 
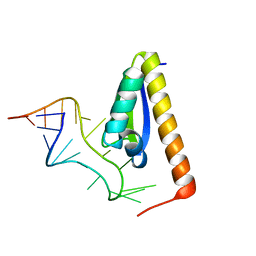 | | CRYSTAL STRUCTURE OF NOVA-2 KH3 K-HOMOLOGY RNA-BINDING DOMAIN BOUND TO 20-MER RNA HAIRPIN | | Descriptor: | 20-MER RNA HAIRPIN, RNA-BINDING PROTEIN NOVA-2 | | Authors: | Lewis, H.A, Musunuru, K, Jensen, K.B, Edo, C, Chen, H. | | Deposit date: | 2000-01-25 | | Release date: | 2000-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Sequence-specific RNA binding by a Nova KH domain: implications for paraneoplastic disease and the fragile X syndrome.
Cell(Cambridge,Mass.), 100, 2000
|
|
1DTJ
 
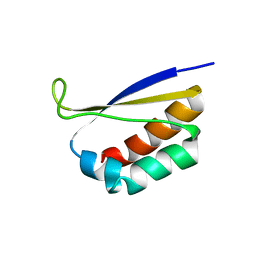 | | CRYSTAL STRUCTURE OF NOVA-2 KH3 K-HOMOLOGY RNA-BINDING DOMAIN | | Descriptor: | RNA-BINDING NEUROONCOLOGICAL VENTRAL ANTIGEN 2 | | Authors: | Lewis, H.A, Chen, H, Edo, C, Buckanovich, R.J, Yang, Y.Y.L, Musunuru, K, Zhong, R, Darnell, R.B, Burley, S.K. | | Deposit date: | 2000-01-12 | | Release date: | 2000-02-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of Nova-1 and Nova-2 K-homology RNA-binding domains.
Structure Fold.Des., 7, 1999
|
|
1DP7
 
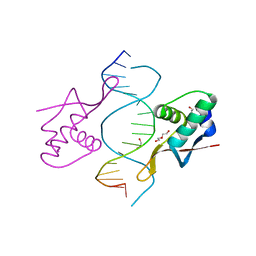 | | COCRYSTAL STRUCTURE OF RFX-DBD IN COMPLEX WITH ITS COGNATE X-BOX BINDING SITE | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*CP*GP*(BRU)P*TP*AP*CP*CP*AP*(BRU)P*GP*GP*TP*AP*AP*CP*G)-3'), ... | | Authors: | Gajiwala, K.S, Chen, H, Cornille, F, Roques, B.P, Reith, W, Mach, B, Burley, S.K. | | Deposit date: | 1999-12-23 | | Release date: | 2000-03-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the winged-helix protein hRFX1 reveals a new mode of DNA binding.
Nature, 403, 2000
|
|
8ZJV
 
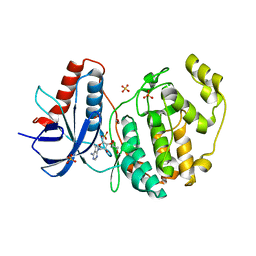 | | Crystal Structure of the ERK2 complexed with 5-Iodotubercidin | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Yang, Y, Fu, L, Chen, R, Cheng, H, Sun, X. | | Deposit date: | 2024-05-15 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the ERK2 complexed with 5-Iodotubercidin
To Be Published
|
|
1DT4
 
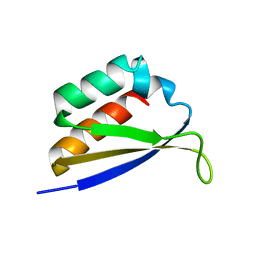 | | CRYSTAL STRUCTURE OF NOVA-1 KH3 K-HOMOLOGY RNA-BINDING DOMAIN | | Descriptor: | NEURO-ONCOLOGICAL VENTRAL ANTIGEN 1 | | Authors: | Lewis, H.A, Chen, H, Edo, C, Buckanovich, R.J, Yang, Y.Y.L. | | Deposit date: | 2000-01-11 | | Release date: | 2000-02-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of Nova-1 and Nova-2 K-homology RNA-binding domains.
Structure Fold.Des., 7, 1999
|
|
1CDW
 
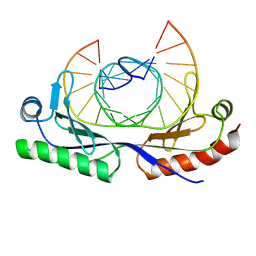 | | HUMAN TBP CORE DOMAIN COMPLEXED WITH DNA | | Descriptor: | DNA (5'-D(*CP*AP*GP*CP*CP*TP*TP*TP*TP*AP*TP*AP*GP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*CP*TP*AP*TP*AP*AP*AP*AP*GP*GP*CP*TP*G)-3'), PROTEIN (TATA BINDING PROTEIN (TBP)) | | Authors: | Nikolov, D.B, Chen, H, Halay, E.D, Hoffmann, A, Roeder, R.G, Burley, S.K. | | Deposit date: | 1996-04-11 | | Release date: | 1996-12-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a human TATA box-binding protein/TATA element complex.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1EDU
 
 | | CRYSTAL STRUCTURE OF THE ENTH DOMAIN OF RAT EPSIN 1 | | Descriptor: | 1,2-ETHANEDIOL, EH domain binding protein EPSIN | | Authors: | Hyman, J.H, Chen, H, Decamilli, P, Brunger, A.T. | | Deposit date: | 2000-01-28 | | Release date: | 2000-05-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Epsin 1 undergoes nucleocytosolic shuttling and its eps15 interactor NH(2)-terminal homology (ENTH) domain, structurally similar to Armadillo and HEAT repeats, interacts with the transcription factor promyelocytic leukemia Zn(2)+ finger protein (PLZF).
J.Cell Biol., 149, 2000
|
|
5CUQ
 
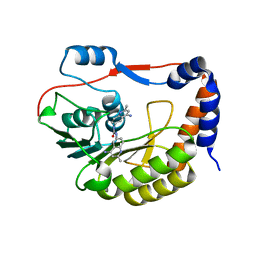 | | Identification and characterization of novel broad spectrum inhibitors of the flavivirus methyltransferase | | Descriptor: | N,N'-BIS(4-AMINO-2-METHYLQUINOLIN-6-YL)UREA, Nonstructural protein NS5 | | Authors: | Brecher, B, Chen, H, Li, Z, Banavali, N.K, Jones, S.A, Zhang, J, Kramer, L.D, Li, H.M. | | Deposit date: | 2015-07-24 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Identification and Characterization of Novel Broad-Spectrum Inhibitors of the Flavivirus Methyltransferase.
Acs Infect Dis., 1, 2015
|
|
4RVM
 
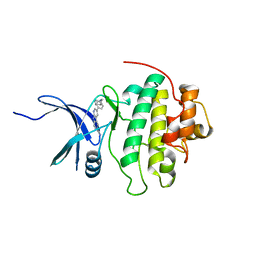 | | CHK1 kinase domain with diazacarbazole compound 19 | | Descriptor: | 3-[4-(piperidin-1-ylmethyl)phenyl]-9H-pyrrolo[2,3-b:5,4-c']dipyridine-6-carbonitrile, Serine/threonine-protein kinase Chk1 | | Authors: | Gazzard, L, Blackwood, E, Burton, B, Chapman, K, Chen, H, Crackett, P, Drobnick, J, Ellwood, C, Epler, J, Flagella, M, Goodacre, S, Halladay, J, Hunt, H, Kintz, S, Lyssikatos, J, MacLeod, C, Ramiscal, S, Schmidt, S, Seward, E, Wiesmann, C, Williams, K, Wu, P, Yee, S, Yen, I, Malek, S. | | Deposit date: | 2014-11-26 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Mitigation of Acetylcholine Esterase Activity in the 1,7-Diazacarbazole Series of Inhibitors of Checkpoint Kinase 1.
J.Med.Chem., 58, 2015
|
|
1T3J
 
 | | Mitofusin domain HR2 V686M/I708M mutant | | Descriptor: | mitofusin 1 | | Authors: | Koshiba, T, Detmer, S.A, Kaiser, J.T, Chen, H, McCaffery, J.M, Chan, D.C. | | Deposit date: | 2004-04-26 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of mitochondrial tethering by mitofusin complexes
Science, 305, 2004
|
|
8EQB
 
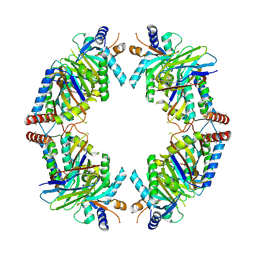 | | FAM46C/BCCIPalpha/Nanobody complex | | Descriptor: | Isoform 2 of BRCA2 and CDKN1A-interacting protein, Synthetic nanobody 1, Terminal nucleotidyltransferase 5C | | Authors: | Liu, S, Chen, H, Yin, Y, Bai, X, Zhang, X. | | Deposit date: | 2022-10-07 | | Release date: | 2023-03-15 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Inhibition of FAM46/TENT5 activity by BCCIP alpha adopting a unique fold.
Sci Adv, 9, 2023
|
|
4R05
 
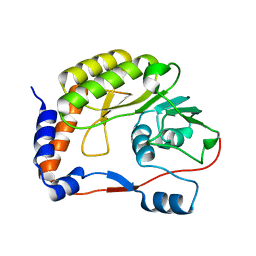 | | Crystal structure of the refolded DENV3 methyltransferase | | Descriptor: | Nonstructural protein NS5 | | Authors: | Brecher, M.B, Li, Z, Zhang, J, Chen, H, Lin, Q, Liu, B, Li, H.M. | | Deposit date: | 2014-07-29 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Refolding of a fully functional flavivirus methyltransferase revealed that S-adenosyl methionine but not S-adenosyl homocysteine is copurified with flavivirus methyltransferase.
Protein Sci., 24, 2015
|
|
2Z6T
 
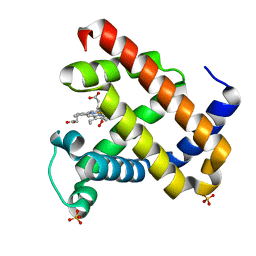 | | Crystal structure of the ferric peroxo myoglobin | | Descriptor: | Myoglobin, PEROXIDE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Unno, M, Kusama, S, Chen, H, Shaik, S, Ikeda-Saito, M. | | Deposit date: | 2007-08-08 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Characterization of the Fleeting Ferric Peroxo Species in Myoglobin: Experiment and Theory
J.Am.Chem.Soc., 129, 2007
|
|
2Z6S
 
 | | Crystal structure of the oxy myoglobin free from X-ray-induced photoreduction | | Descriptor: | Myoglobin, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Unno, M, Kusama, S, Chen, H, Shaik, S, Ikeda-Saito, M. | | Deposit date: | 2007-08-08 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Characterization of the Fleeting Ferric Peroxo Species in Myoglobin: Experiment and Theory
J.Am.Chem.Soc., 129, 2007
|
|
3Q2C
 
 | | Binding properties to HLA class I molecules and the structure of the leukocyte Ig-like receptor A3 (LILRA3/ILT6/LIR4/CD85e) | | Descriptor: | Leukocyte immunoglobulin-like receptor subfamily A member 3 | | Authors: | Ryu, M, Chen, Y, Qi, J.X, Liu, J, Shi, Y, Cheng, H, Gao, G.F. | | Deposit date: | 2010-12-20 | | Release date: | 2011-07-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | LILRA3 binds both classical and non-classical HLA class I molecules but with reduced affinities compared to LILRB1/LILRB2: structural evidence
Plos One, 6, 2011
|
|
5AD2
 
 | | Bivalent binding to BET bromodomains | | Descriptor: | (3R)-4-(2-{4-[1-(3-chloro[1,2,4]triazolo[4,3-b]pyridazin-6-yl)-4-piperidinyl]phenoxy}ethyl)-1,3-dimethyl-2-piperazinone, BROMODOMAIN-CONTAINING PROTEIN 4 | | Authors: | Waring, M.J, Chen, H, Rabow, A.A, Walker, G, Bobby, R, Boiko, S, Bradbury, R.H, Callis, R, Dale, I, Daniels, D, Flavell, L, Holdgate, G, Jowitt, T.A, Kikhney, A, McAlister, M, Ogg, D, Patel, J, Petteruti, P, Robb, G.R, Robers, M, Stratton, N, Svergun, D.I, Wang, W, Whittaker, D. | | Deposit date: | 2015-08-19 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Potent and Selective Bivalent Inhibitors of Bet Bromodomains
Nat.Chem.Biol., 12, 2016
|
|
5AD3
 
 | | Bivalent binding to BET bromodomains | | Descriptor: | 3-methoxy-N-[2-[4-[1-(3-methoxy-[1,2,4]triazolo[4,3-b]pyridazin-6-yl)-4-piperidyl]phenoxy]ethyl]-N-methyl-[1,2,4]triazolo[4,3-b]pyridazin-6-amine, BROMODOMAIN-CONTAINING PROTEIN 4 | | Authors: | Waring, M.J, Chen, H, Rabow, A.A, Walker, G, Bobby, R, Boiko, S, Bradbury, R.H, Callis, R, Dale, I, Daniels, D, Flavell, L, Holdgate, G, Jowitt, T.A, Kikhney, A, McAlister, M, Ogg, D, Patel, J, Petteruti, P, Robb, G.R, Robers, M, Stratton, N, Svergun, D.I, Wang, W, Whittaker, D. | | Deposit date: | 2015-08-19 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Potent and Selective Bivalent Inhibitors of Bet Bromodomains
Nat.Chem.Biol., 12, 2016
|
|
5CFB
 
 | | Crystal Structure of Human Glycine Receptor alpha-3 Bound to Strychnine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alpha-3,Glycine receptor subunit alpha-3, ... | | Authors: | Shaffer, P.L, Huang, X, Chen, H. | | Deposit date: | 2015-07-08 | | Release date: | 2015-09-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Crystal structure of human glycine receptor-alpha 3 bound to antagonist strychnine.
Nature, 526, 2015
|
|
5VDI
 
 | | Crystal Structure of Human Glycine Receptor alpha-3 Mutant N38Q Bound to AM-3607, Glycine, and Ivermectin | | Descriptor: | (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17 -oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H-spiro[11,15-methanofuro[4,3,2-pq][2,6]benzodioxacy clooctadecine-13,2'-pyran]-7-yl 2,6-dideoxy-4-O-(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)-3-O-methyl-alpha-L-arabino-hexopyranoside, (3S,3aS,9bS)-2-[(2H-1,3-benzodioxol-5-yl)sulfonyl]-3,5-dimethyl-1,2,3,3a,5,9b-hexahydro-4H-pyrrolo[3,4-c][1,6]naphthyridin-4-one, GLYCINE, ... | | Authors: | Shaffer, P.L, Huang, X, Chen, H. | | Deposit date: | 2017-04-03 | | Release date: | 2017-05-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structures of Human GlyR alpha 3 Bound to Ivermectin.
Structure, 25, 2017
|
|
5VDH
 
 | | Crystal Structure of Human Glycine Receptor alpha-3 Bound to AM-3607, Glycine, and Ivermectin | | Descriptor: | (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17 -oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H-spiro[11,15-methanofuro[4,3,2-pq][2,6]benzodioxacy clooctadecine-13,2'-pyran]-7-yl 2,6-dideoxy-4-O-(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)-3-O-methyl-alpha-L-arabino-hexopyranoside, (3S,3aS,9bS)-2-[(2H-1,3-benzodioxol-5-yl)sulfonyl]-3,5-dimethyl-1,2,3,3a,5,9b-hexahydro-4H-pyrrolo[3,4-c][1,6]naphthyridin-4-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shaffer, P.L, Huang, X, Chen, H. | | Deposit date: | 2017-04-03 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structures of Human GlyR alpha 3 Bound to Ivermectin.
Structure, 25, 2017
|
|
2HPV
 
 | | Crystal structure of FMN-Dependent azoreductase from Enterococcus faecalis | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase | | Authors: | Liu, Z.J, Chen, L, Chen, H, Rose, J, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-07-17 | | Release date: | 2006-09-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Fmn-Dependent Azoreductase from Enterococcus faecalis at 2.00 A resolution
To be Published
|
|
5TIO
 
 | | Crystal Structure of Human Glycine Receptor alpha-3 Bound to AM-3607 | | Descriptor: | (3S,3aS,9bS)-2-[(2H-1,3-benzodioxol-5-yl)sulfonyl]-3,5-dimethyl-1,2,3,3a,5,9b-hexahydro-4H-pyrrolo[3,4-c][1,6]naphthyridin-4-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | Authors: | Shaffer, P.L, Huang, X, Chen, H. | | Deposit date: | 2016-10-03 | | Release date: | 2017-01-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal Structures of Human GlyRa3 Bound to a Novel Class of Potentiators with Efficacy in a Mouse Model of Neuropathic Pain
To Be Published
|
|
5TIN
 
 | | Crystal Structure of Human Glycine Receptor alpha-3 Mutant N38Q Bound to AM-3607 | | Descriptor: | (3S,3aS,9bS)-2-[(2H-1,3-benzodioxol-5-yl)sulfonyl]-3,5-dimethyl-1,2,3,3a,5,9b-hexahydro-4H-pyrrolo[3,4-c][1,6]naphthyridin-4-one, CHLORIDE ION, GLYCINE, ... | | Authors: | Shaffer, P.L, Huang, X, Chen, H. | | Deposit date: | 2016-10-03 | | Release date: | 2017-01-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal Structures of Human GlyRa3 Bound to a Novel Class of Potentiators with Efficacy in a Mouse Model of Neuropathic Pain
To Be Published
|
|
