1TVW
 
 | |
1TSZ
 
 | | THYMIDYLATE SYNTHASE R179K MUTANT | | Descriptor: | PHOSPHATE ION, THYMIDYLATE SYNTHASE | | Authors: | Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1995-12-05 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Contribution of a salt bridge to binding affinity and dUMP orientation to catalytic rate: mutation of a substrate-binding arginine in thymidylate synthase.
Protein Eng., 9, 1996
|
|
1KCE
 
 | | E. COLI THYMIDYLATE SYNTHASE MUTANT E58Q IN COMPLEX WITH CB3717 AND 2'-DEOXYURIDINE 5'-MONOPHOSPHATE (DUMP) | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Sage, C.R, Rutenber, E.E, Stout, T.J, Stroud, R.M. | | Deposit date: | 1996-10-22 | | Release date: | 1997-04-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An essential role for water in an enzyme reaction mechanism: the crystal structure of the thymidylate synthase mutant E58Q.
Biochemistry, 35, 1996
|
|
1JTU
 
 | | E. coli Thymidylate Synthase in a Complex with dUMP and LY338913, A Polyglutamylated Pyrrolo(2,3-d)pyrimidine-based Antifolate | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-{4-[4-(4-{4-[2-(2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDIN-5-YL)-ETHYL]-BENZOYLAMINO}-4-CARBOXY-BUTYRYLAMIN O)-4-CARBOXY-BUTYRYLAMINO}-PENTANEDIOIC ACID, THYMIDYLATE SYNTHASE | | Authors: | Sayre, P.H, Finer-Moore, J.S, Fritz, T.A, Biermann, D, Gates, S.B, MacKellar, W.C, Patel, V.F, Stroud, R.M. | | Deposit date: | 2001-08-22 | | Release date: | 2001-09-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multi-targeted antifolates aimed at avoiding drug resistance form covalent closed inhibitory complexes with human and Escherichia coli thymidylate synthases.
J.Mol.Biol., 313, 2001
|
|
1BJG
 
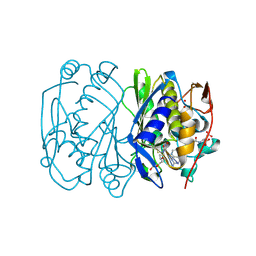 | | D221(169)N MUTANT DOES NOT PROMOTE OPENING OF THE COFACTOR IMIDAZOLIDINE RING | | Descriptor: | 5,10-METHYLENE-6-HYDROFOLIC ACID, 5-FLUORO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Sage, C.R, Michelitsch, M.D, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1998-06-25 | | Release date: | 1998-11-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | D221 in thymidylate synthase controls conformation change, and thereby opening of the imidazolidine.
Biochemistry, 37, 1998
|
|
1BO7
 
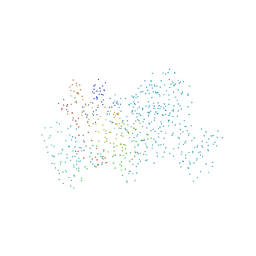 | | THYMIDYLATE SYNTHASE R179T MUTANT | | Descriptor: | THYMIDYLATE SYNTHASE, URIDINE-5'-MONOPHOSPHATE | | Authors: | Morse, R, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1998-08-10 | | Release date: | 1998-08-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Energetic contributions of four arginines to phosphate-binding in thymidylate synthase are more than additive and depend on optimization of "effective charge balance".
Biochemistry, 39, 2000
|
|
1BPJ
 
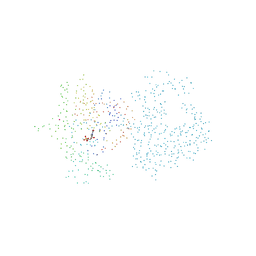 | | THYMIDYLATE SYNTHASE R178T, R179T DOUBLE MUTANT | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, POTASSIUM ION, PROTEIN (THYMIDYLATE SYNTHASE) | | Authors: | Morse, R.J, Finer-Moore, J.S, Stroud, R.M. | | Deposit date: | 1998-08-11 | | Release date: | 1998-08-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Energetic contributions of four arginines to phosphate-binding in thymidylate synthase are more than additive and depend on optimization of "effective charge balance".
Biochemistry, 39, 2000
|
|
1BP0
 
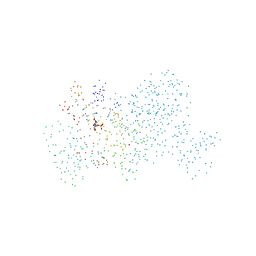 | | THYMIDYLATE SYNTHASE R23I MUTANT | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, POTASSIUM ION, PROTEIN (THYMIDYLATE SYNTHASE) | | Authors: | Morse, R, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1998-08-11 | | Release date: | 2000-01-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Energetic contributions of four arginines to phosphate-binding in thymidylate synthase are more than additive and depend on optimization of "effective charge balance".
Biochemistry, 39, 2000
|
|
1BP6
 
 | | THYMIDYLATE SYNTHASE R23I, R179T DOUBLE MUTANT | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, POTASSIUM ION, PROTEIN (THYMIDYLATE SYNTHASE) | | Authors: | Morse, R.J, Finer-Moore, J.S, Stroud, R.M. | | Deposit date: | 1998-08-13 | | Release date: | 1998-08-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Energetic contributions of four arginines to phosphate-binding in thymidylate synthase are more than additive and depend on optimization of "effective charge balance".
Biochemistry, 39, 2000
|
|
1BO8
 
 | | THYMIDYLATE SYNTHASE R178T MUTANT | | Descriptor: | POTASSIUM ION, THYMIDYLATE SYNTHASE, URIDINE-5'-MONOPHOSPHATE | | Authors: | Morse, R, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1998-08-10 | | Release date: | 1998-08-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Energetic contributions of four arginines to phosphate-binding in thymidylate synthase are more than additive and depend on optimization of "effective charge balance".
Biochemistry, 39, 2000
|
|
8SX9
 
 | |
8SX8
 
 | |
8SX7
 
 | |
8SXB
 
 | |
8SWN
 
 | |
8SXA
 
 | |
2TDM
 
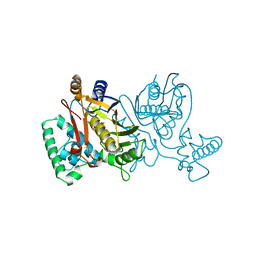 | | STRUCTURE OF THYMIDYLATE SYNTHASE | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1997-03-28 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Refined structures of substrate-bound and phosphate-bound thymidylate synthase from Lactobacillus casei.
J.Mol.Biol., 232, 1993
|
|
2TDD
 
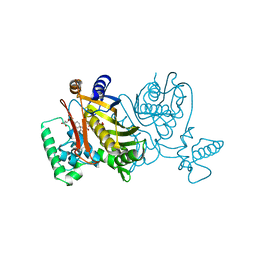 | | STRUCTURES OF THYMIDYLATE SYNTHASE WITH A C-TERMINAL DELETION: ROLE OF THE C-TERMINUS IN ALIGNMENT OF D/UMP AND CH2H4FOLATE | | Descriptor: | 5-FLUORO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, 5-HYDROXYMETHYLENE-6-HYDROFOLIC ACID, THYMIDYLATE SYNTHASE | | Authors: | Perry, K.M, Carreras, C.W, Chang, L.C, Santi, D.V, Stroud, R.M. | | Deposit date: | 1993-04-05 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of thymidylate synthase with a C-terminal deletion: role of the C-terminus in alignment of 2'-deoxyuridine 5'-monophosphate and 5,10-methylenetetrahydrofolate.
Biochemistry, 32, 1993
|
|
7TDF
 
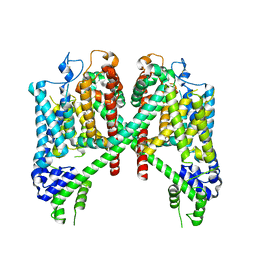 | | AtTPC1 D454N with 1 mM EDTA state I | | Descriptor: | Two pore calcium channel protein 1 | | Authors: | Dickinson, M.S, Stroud, R.M. | | Deposit date: | 2021-12-31 | | Release date: | 2022-02-02 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Molecular basis of multistep voltage activation in plant two-pore channel 1.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7TDD
 
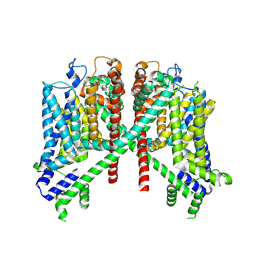 | | AtTPC1 D454N-EDTA state II | | Descriptor: | Two pore calcium channel protein 1 | | Authors: | Dickinson, M.S, Stroud, R.M. | | Deposit date: | 2021-12-30 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis of multistep voltage activation in plant two-pore channel 1.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7TDE
 
 | | AtTPC1 DDE mutant with 1 mM Ca2+ | | Descriptor: | Two pore calcium channel protein 1 | | Authors: | Dickinson, M.S, Stroud, R.M. | | Deposit date: | 2021-12-31 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis of multistep voltage activation in plant two-pore channel 1.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7T3N
 
 | | R184Q/E191Q mutant of rat vesicular glutamate transporter 2 (VGLUT2) | | Descriptor: | (1S,3R)-1-AMINOCYCLOPENTANE-1,3-DICARBOXYLIC ACID, (2R)-2-(methoxymethyl)-4-{[(25R)-spirost-5-en-3beta-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, Vesicular glutamate transporter 2 | | Authors: | Li, F, Stroud, R.M, Edwards, R.H. | | Deposit date: | 2021-12-08 | | Release date: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Substrate Recognition, Directional Flux, and Allostery in a Synaptic Vesicle Glutamate Transporter
To Be Published
|
|
7T3O
 
 | |
3P5S
 
 | | Structural insights into the catalytic mechanism of CD38: Evidence for a conformationally flexible covalent enzyme-substrate complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CD38 molecule, SULFATE ION, ... | | Authors: | Egea, P.F, Muller-Stauffler, H, Kohn, I, Cakou-Kefir, C, Stroud, R.M, Kellenberburger, E, Schuber, F. | | Deposit date: | 2010-10-10 | | Release date: | 2011-10-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Insights into the mechanism of bovine CD38/NAD+glycohydrolase from the X-ray structures of its Michaelis complex and covalently-trapped intermediates.
Plos One, 7, 2012
|
|
3R1F
 
 | | Crystal structure of a key regulator of virulence in Mycobacterium tuberculosis | | Descriptor: | ESX-1 secretion-associated regulator EspR | | Authors: | Rosenberg, O.S, Dovey, C, Finer-Moore, J, Stroud, R.M, Cox, J.S. | | Deposit date: | 2011-03-10 | | Release date: | 2011-08-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | EspR, a key regulator of Mycobacterium tuberculosis virulence, adopts a unique dimeric structure among helix-turn-helix proteins.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
