2F6F
 
 | | The structure of the S295F mutant of human PTP1B | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Tyrosine-protein phosphatase, ... | | Authors: | Montalibet, J, Skorey, K, McKay, D, Scapin, G, Asante-Appiah, E, Kennedy, B.P. | | Deposit date: | 2005-11-29 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Residues distant from the active site influence protein-tyrosine phosphatase 1B inhibitor binding.
J.Biol.Chem., 281, 2006
|
|
2I0H
 
 | | The structure of p38alpha in complex with an arylpyridazinone | | Descriptor: | 2-(3-{(2-CHLORO-4-FLUOROPHENYL)[1-(2-CHLOROPHENYL)-6-OXO-1,6-DIHYDROPYRIDAZIN-3-YL]AMINO}PROPYL)-1H-ISOINDOLE-1,3(2H)-DIONE, GLYCEROL, Mitogen-activated protein kinase 14 | | Authors: | Natarajan, S.R, Heller, S.T, Nam, K, Singh, S.B, Scapin, G, Patel, S, Thompson, J.E, Fitzgerald, C.E, O'Keefe, S.J. | | Deposit date: | 2006-08-10 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | p38 MAP Kinase Inhibitors Part 6: 2-Arylpyridazin-3-ones as templates for inhibitor design.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1F06
 
 | | THREE DIMENSIONAL STRUCTURE OF THE TERNARY COMPLEX OF CORYNEBACTERIUM GLUTAMICUM DIAMINOPIMELATE DEHYDROGENASE NADPH-L-2-AMINO-6-METHYLENE-PIMELATE | | Descriptor: | L-2-AMINO-6-METHYLENE-PIMELIC ACID, MESO-DIAMINOPIMELATE D-DEHYDROGENASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cirilli, M, Scapin, G, Sutherland, A, Caplan, J.F, Vederas, J.C, Blanchard, J.S. | | Deposit date: | 2000-05-14 | | Release date: | 2001-05-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The three-dimensional structure of the ternary complex of Corynebacterium glutamicum diaminopimelate dehydrogenase-NADPH-L-2-amino-6-methylene-pimelate.
Protein Sci., 9, 2000
|
|
2FJM
 
 | | The structure of phosphotyrosine phosphatase 1B in complex with compound 2 | | Descriptor: | (4-{(2S,4E)-2-(1H-1,2,3-BENZOTRIAZOL-1-YL)-2-[4-(METHOXYCARBONYL)PHENYL]-5-PHENYLPENT-4-ENYL}PHENYL)(DIFLUORO)METHYLPHOSPHONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Asante-Appiah, E, Patel, S, Desponts, C, Taylor, J.M, Lau, C, Dufresne, C, Therien, M, Friesen, R, Becker, J.W, Leblanc, Y, Scapin, G. | | Deposit date: | 2006-01-03 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformation-assisted inhibition of protein-tyrosine phosphatase-1B elicits inhibitor selectivity over T-cell protein-tyrosine phosphatase.
J.Biol.Chem., 281, 2006
|
|
2FJN
 
 | | The structure of phosphotyrosine phosphatase 1B in complex with compound 2 | | Descriptor: | (4-{(2S,4E)-2-(1H-1,2,3-BENZOTRIAZOL-1-YL)-2-[4-(METHOXYCARBONYL)PHENYL]-5-PHENYLPENT-4-ENYL}PHENYL)(DIFLUORO)METHYLPHOSPHONIC ACID, CHLORIDE ION, Tyrosine-protein phosphatase, ... | | Authors: | Asante-Appiah, E, Patel, S, Desponts, C, Taylor, J.M, Lau, C, Dufresne, C, Therien, M, Friesen, R, Becker, J.W, Leblanc, Y, Kennedy, B.P, Scapin, G. | | Deposit date: | 2006-01-03 | | Release date: | 2006-01-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformation-assisted inhibition of protein-tyrosine phosphatase-1B elicits inhibitor selectivity over T-cell protein-tyrosine phosphatase.
J.Biol.Chem., 281, 2006
|
|
1B9H
 
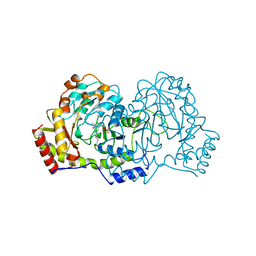 | | CRYSTAL STRUCTURE OF 3-AMINO-5-HYDROXYBENZOIC ACID (AHBA) SYNTHASE | | Descriptor: | PROTEIN (3-AMINO-5-HYDROXYBENZOIC ACID SYNTHASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Eads, J.C, Beeby, M, Scapin, G, Yu, T.-W, Floss, H.G. | | Deposit date: | 1999-02-11 | | Release date: | 1999-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of 3-amino-5-hydroxybenzoic acid (AHBA) synthase.
Biochemistry, 38, 1999
|
|
1BWZ
 
 | |
1C3V
 
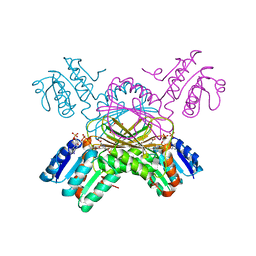 | | DIHYDRODIPICOLINATE REDUCTASE FROM MYCOBACTERIUM TUBERCULOSIS COMPLEXED WITH NADPH AND PDC | | Descriptor: | DIHYDRODIPICOLINATE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PYRIDINE-2,6-DICARBOXYLIC ACID, ... | | Authors: | Cirilli, M, Zheng, R, Scapin, G, Blanchard, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 1999-07-28 | | Release date: | 2003-08-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | The three-dimensional structures of the Mycobacterium tuberculosis
dihydrodipicolinate reductase-NADH-2,6-PDC and -NADPH-2,6-PDC complexes.
Structural and mutagenic analysis of relaxed nucleotide specificity
Biochemistry, 42, 2003
|
|
1B9I
 
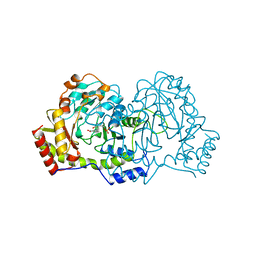 | | CRYSTAL STRUCTURE OF 3-AMINO-5-HYDROXYBENZOIC ACID (AHBA) SYNTHASE | | Descriptor: | 3-[O-PHOSPHONOPYRIDOXYL]--AMINO-BENZOIC ACID, PROTEIN (3-AMINO-5-HYDROXYBENZOIC ACID SYNTHASE) | | Authors: | Eads, J.C, Beeby, M, Scapin, G, Yu, T.-W, Floss, H.G. | | Deposit date: | 1999-02-11 | | Release date: | 1999-08-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of 3-amino-5-hydroxybenzoic acid (AHBA) synthase.
Biochemistry, 38, 1999
|
|
1M7Q
 
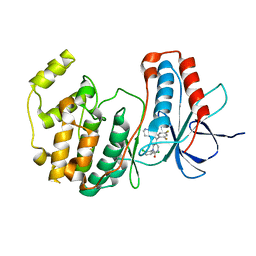 | | Crystal structure of p38 MAP kinase in complex with a dihydroquinazolinone inhibitor | | Descriptor: | 1-(2,6-DICHLOROPHENYL)-5-(2,4-DIFLUOROPHENYL)-7-PIPERAZIN-1-YL-3,4-DIHYDROQUINAZOLIN-2(1H)-ONE, Mitogen-activated protein kinase 14, SULFATE ION | | Authors: | Stelmach, J.E, Liu, L, Patel, S.B, Pivnichny, J.V, Scapin, G, Singh, S, Hop, C.E.C.A, Wang, Z, Cameron, P.M, Nichols, E.A, O'Keefe, S.J, O'Neill, E.A, Schmatz, D.M, Schwartz, C.D, Thompson, C.M, Zaller, D.M, Doherty, J.B. | | Deposit date: | 2002-07-22 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and synthesis of potent, orally bioavailable dihydroquinazolinone inhibitors of p38 MAP kinase.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
1PA1
 
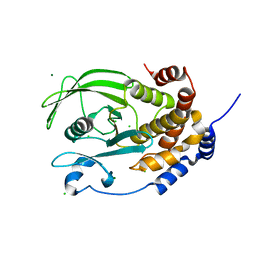 | | Crystal structure of the C215D mutant of protein tyrosine phosphatase 1B | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Protein-tyrosine phosphatase, ... | | Authors: | Romsicki, Y, Scapin, G, Beaulieu-Audy, V, Patel, S.B, Becker, J.W, Kennedy, B, Asante-Appiah, E. | | Deposit date: | 2003-05-13 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Functional characterization and crystal structure of the C215D mutant of protein-tyrosine phosphatase-1B
J.Biol.Chem., 278, 2003
|
|
1OUK
 
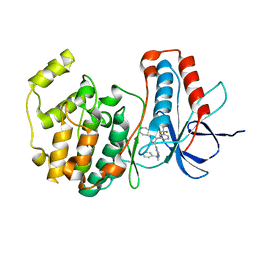 | | The structure of p38 alpha in complex with a pyridinylimidazole inhibitor | | Descriptor: | 4-[5-[2-(1-PHENYL-ETHYLAMINO)-PYRIMIDIN-4-YL]-1-METHYL-4-(3-TRIFLUOROMETHYLPHENYL)-1H-IMIDAZOL-2-YL]-PIPERIDINE, Mitogen-activated protein kinase 14, SULFATE ION | | Authors: | Fitzgerald, C.E, Patel, S.B, Becker, J.W, Cameron, P.M, Zaller, D, Pikounis, V.B, O'Keefe, S.J, Scapin, G. | | Deposit date: | 2003-03-24 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for p38alpha MAP kinase quinazolinone and pyridol-pyrimidine inhibitor specificity
Nat.Struct.Biol., 10, 2003
|
|
1OUY
 
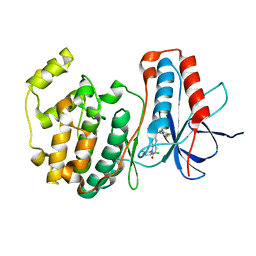 | | The structure of p38 alpha in complex with a dihydropyrido-pyrimidine inhibitor | | Descriptor: | 1-(2,6-DICHLOROPHENYL)-6-[(2,4-DIFLUOROPHENYL)SULFANYL]-7-(1,2,3,6-TETRAHYDRO-4-PYRIDINYL)-3,4-DIHYDROPYRIDO[3,2-D]PYRIMIDIN-2(1H)-ONE, Mitogen-activated protein kinase 14 | | Authors: | Fitzgerald, C.E, Patel, S.B, Becker, J.W, Cameron, P.M, Zaller, D, Pikounis, V.B, O'Keefe, S.J, Scapin, G. | | Deposit date: | 2003-03-25 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for p38alpha MAP kinase quinazolinone and pyridol-pyrimidine inhibitor specificity
Nat.Struct.Biol., 10, 2003
|
|
1MAS
 
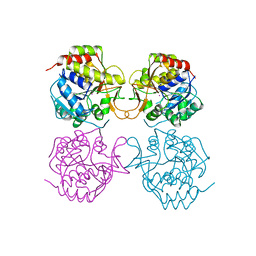 | | PURINE NUCLEOSIDE HYDROLASE | | Descriptor: | INOSINE-URIDINE NUCLEOSIDE N-RIBOHYDROLASE, POTASSIUM ION | | Authors: | Degano, M, Gopaul, D.N, Scapin, G, Schramm, V.L, Sacchettini, J.C. | | Deposit date: | 1995-12-18 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structure of the inosine-uridine nucleoside N-ribohydrolase from Crithidia fasciculata.
Biochemistry, 35, 1996
|
|
1P9L
 
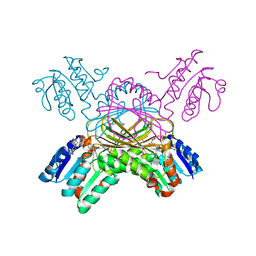 | | Structure of M. tuberculosis dihydrodipicolinate reductase in complex with NADH and 2,6 PDC | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, PYRIDINE-2,6-DICARBOXYLIC ACID, TETRAETHYLENE GLYCOL, ... | | Authors: | Cirilli, M, Zheng, R, Scapin, G, Blanchard, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-05-12 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The three-dimensional structures of the Mycobacterium tuberculosis dihydrodipicolinate reductase-NADH-2,6-PDC and -NADPH-2,6-PDC complexes. Structural and mutagenic analysis of relaxed nucleotide specificity.
Biochemistry, 42, 2003
|
|
7UYA
 
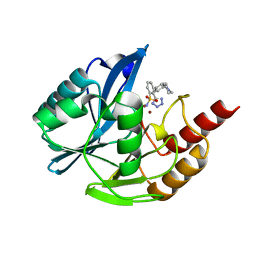 | | Inhibitor bound VIM1 | | Descriptor: | (2M)-4'-(piperidin-4-yl)-2-(1H-tetrazol-5-yl)[1,1'-biphenyl]-3-sulfonamide, Beta-lactamase VIM-1, ZINC ION | | Authors: | Fischmann, T.O, Scapin, G. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Structure Guided Discovery of Novel Pan Metallo-beta-Lactamase Inhibitors with Improved Gram-Negative Bacterial Cell Penetration.
J.Med.Chem., 67, 2024
|
|
7UYD
 
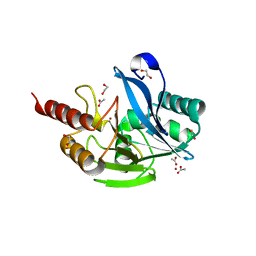 | | Inhibitor bound VIM1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Metallo-beta-lactamase VIM-2-like protein, ... | | Authors: | Fischmann, T.O, Scapin, G. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure Guided Discovery of Novel Pan Metallo-beta-Lactamase Inhibitors with Improved Gram-Negative Bacterial Cell Penetration.
J.Med.Chem., 67, 2024
|
|
7UYB
 
 | | Inhibitor bound VIM1 | | Descriptor: | (2M)-4'-(piperidin-4-yl)-2-(1H-tetrazol-5-yl)-4-(trifluoromethyl)[1,1'-biphenyl]-3-sulfonamide, Beta-lactamase VIM-1, ZINC ION | | Authors: | Fischmann, T.O, Scapin, G. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Structure Guided Discovery of Novel Pan Metallo-beta-Lactamase Inhibitors with Improved Gram-Negative Bacterial Cell Penetration.
J.Med.Chem., 67, 2024
|
|
7UYC
 
 | | Inhibitor bound VIM1 | | Descriptor: | (2P)-4'-(piperidin-4-yl)-4-[(piperidin-4-yl)methyl]-2-(1H-tetrazol-5-yl)[1,1'-biphenyl]-3-sulfonamide, Beta-lactamase VIM-1, MAGNESIUM ION, ... | | Authors: | Fischmann, T.O, Scapin, G. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Structure Guided Discovery of Novel Pan Metallo-beta-Lactamase Inhibitors with Improved Gram-Negative Bacterial Cell Penetration.
J.Med.Chem., 67, 2024
|
|
1TW4
 
 | | Crystal Structure of Chicken Liver Basic Fatty Acid Binding Protein (Bile Acid Binding Protein) Complexed With Cholic Acid | | Descriptor: | CHOLIC ACID, Fatty acid-binding protein | | Authors: | Nichesola, D, Perduca, M, Capaldi, S, Carrizo, M.E, Righetti, P.G, Monaco, H.L. | | Deposit date: | 2004-06-30 | | Release date: | 2004-11-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of chicken liver basic Fatty Acid-binding protein complexed with cholic acid
Biochemistry, 43, 2004
|
|
1TVQ
 
 | | Crystal Structure of Apo Chicken Liver Basic Fatty Acid Binding Protein (or Bile Acid Binding Protein) | | Descriptor: | Fatty acid-binding protein | | Authors: | Nichesola, D, Perduca, M, Capaldi, S, Carrizo, M.E, Righetti, P.G, Monaco, H.L. | | Deposit date: | 2004-06-30 | | Release date: | 2004-11-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of chicken liver basic Fatty Acid-binding protein complexed with cholic acid
Biochemistry, 43, 2004
|
|
8GK7
 
 | | MsbA bound to cerastecin C | | Descriptor: | 2-[(4-butylbenzene-1-sulfonyl)amino]-5-[(3-{4-[(4-butylbenzene-1-sulfonyl)amino]-3-carboxyanilino}-3-oxopropyl)carbamoyl]benzoic acid, Lipid A export ATP-binding/permease protein MsbA, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Chen, Y, Klein, D. | | Deposit date: | 2023-03-17 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Cerastecins inhibit membrane lipooligosaccharide transport in drug-resistant Acinetobacter baumannii.
Nat Microbiol, 9, 2024
|
|
1G5W
 
 | | SOLUTION STRUCTURE OF HUMAN HEART-TYPE FATTY ACID BINDING PROTEIN | | Descriptor: | FATTY ACID-BINDING PROTEIN | | Authors: | Luecke, C, Rademacher, M, Zimmerman, A, van Moerkerk, H.T.B, Veerkamp, J.H, Rueterjans, H. | | Deposit date: | 2000-11-02 | | Release date: | 2001-03-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Spin-system heterogeneities indicate a selected-fit mechanism in fatty acid binding to heart-type fatty acid-binding protein (H-FABP).
Biochem.J., 354, 2001
|
|
9CSG
 
 | | Human Serum Albumin Bound to Cerastecin Compound 5e | | Descriptor: | 2-(4-butylbenzene-1-sulfonamido)-5-(4-{3-carboxy-4-[4-(2-methoxyethyl)benzene-1-sulfonamido]anilino}-4-oxobutanamido)benzoic acid, Albumin, MYRISTIC ACID | | Authors: | Hruza, A, Klein, D.J, Ishchenko, A. | | Deposit date: | 2024-07-23 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.914 Å) | | Cite: | Cerastecin Inhibition of the Lipooligosaccharide Transporter MsbA to Combat Acinetobacter baumannii : From Screening Impurity to In Vivo Efficacy.
J.Med.Chem., 67, 2024
|
|
9CSI
 
 | | A. baumannii MsbA Bound to Cerastecin Compound 5 | | Descriptor: | 3,3'-[(1,4-dioxobutane-1,4-diyl)bis(azanediyl)]bis[(4-butylbenzene-1-sulfonamido)benzoic acid], Lipid A export ATP-binding/permease protein MsbA, MAGNESIUM ION, ... | | Authors: | Klein, D.J, Ishchenko, A, Soisson, S, Cheng, R, Hennig, M. | | Deposit date: | 2024-07-23 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Cerastecin Inhibition of the Lipooligosaccharide Transporter MsbA to Combat Acinetobacter baumannii : From Screening Impurity to In Vivo Efficacy.
J.Med.Chem., 67, 2024
|
|
