5MCQ
 
 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH ACTIVE SITE AND EXOSITE BINDING PEPTIDE INHIBITOR | | Descriptor: | 1,2-ETHANEDIOL, BACE-1 ACTIVE AND EXOSITE BINDING INHIBITOR, Beta-secretase 1 | | Authors: | Kuglstatter, A, Stihle, M, Benz, J. | | Deposit date: | 2016-11-10 | | Release date: | 2017-09-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Potent and Selective BACE-1 Peptide Inhibitors Lower Brain A beta Levels Mediated by Brain Shuttle Transport.
EBioMedicine, 24, 2017
|
|
8TED
 
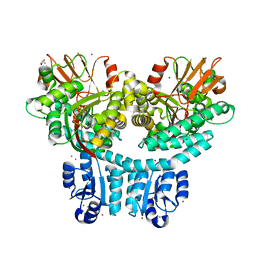 | |
1TD0
 
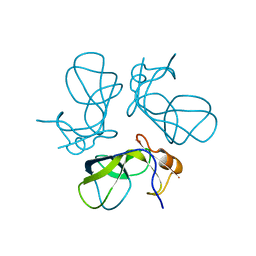 | | Viral capsid protein SHP at pH 5.5 | | Descriptor: | Head decoration protein | | Authors: | Chang, C, Forrer, P, Ott, D, Wlodawer, A, Plueckthun, A. | | Deposit date: | 2004-05-21 | | Release date: | 2004-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Kinetic Stability and Crystal Structure of the Viral Capsid Protein SHP
J.Mol.Biol., 344, 2004
|
|
4CMS
 
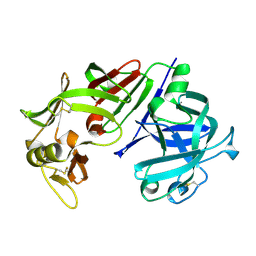 | | X-RAY ANALYSES OF ASPARTIC PROTEINASES IV. STRUCTURE AND REFINEMENT AT 2.2 ANGSTROMS RESOLUTION OF BOVINE CHYMOSIN | | Descriptor: | CHYMOSIN B | | Authors: | Newman, M, Frazao, C, Khan, G, Tickle, I.J, Blundell, T.L, Safro, M, Andreeva, N, Zdanov, A. | | Deposit date: | 1991-11-01 | | Release date: | 1991-11-07 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray analyses of aspartic proteinases. IV. Structure and refinement at 2.2 A resolution of bovine chymosin.
J.Mol.Biol., 221, 1991
|
|
1TDW
 
 | | Crystal structure of double truncated human phenylalanine hydroxylase BH4-responsive PKU mutant A313T. | | Descriptor: | FE (III) ION, Phenylalanine-4-hydroxylase | | Authors: | Erlandsen, H, Pey, A.L, Gamez, A, Perez, B, Desviat, L.R, Aguado, C, Koch, R, Surendran, S, Tyring, S, Matalon, R, Scriver, C.R, Ugarte, M, Martinez, A, Stevens, R.C. | | Deposit date: | 2004-05-24 | | Release date: | 2004-11-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Correction of kinetic and stability defects by tetrahydrobiopterin in phenylketonuria patients with certain phenylalanine hydroxylase mutations.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
8TEF
 
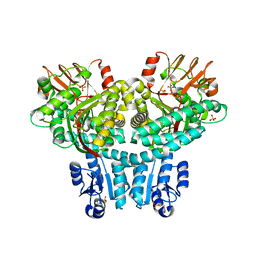 | |
6RUH
 
 | |
5MDK
 
 | | Crystal structure of an O2-tolerant [NiFe]-hydrogenase from Ralstonia eutropha in its as-isolated form (oxidized state - state 3) | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, FE4-S3 CLUSTER, ... | | Authors: | Schmidt, A, Kalms, J, Scheerer, P. | | Deposit date: | 2016-11-11 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Tracking the route of molecular oxygen in O2-tolerant membrane-bound [NiFe] hydrogenase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8T9A
 
 | | CryoEM structure of human DDB1-DCAF12 in complex with MAGEA3 | | Descriptor: | DDB1- and CUL4-associated factor 12, DNA damage-binding protein 1, Melanoma-associated antigen 3 | | Authors: | Duda, D, Righetto, G, Li, Y, Loppnau, P, Seitova, A, Santhakumar, V, Halabelian, L, Yin, Y. | | Deposit date: | 2023-06-23 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | CryoEM structure of human DDB1-DCAF12 in complex with MAGEA3
To Be Published
|
|
6RT0
 
 | | cryo-em structure of alpha-synuclein fibril polymorph 2A | | Descriptor: | Alpha-synuclein | | Authors: | Guerrero-Ferreira, R, Taylor, N.M.I, Arteni, A.A, Melki, R, Meier, B.H, Bockmann, A, Bousset, L, Stahlberg, H. | | Deposit date: | 2019-05-22 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Two new polymorphic structures of human full-length alpha-synuclein fibrils solved by cryo-electron microscopy.
Elife, 8, 2019
|
|
6RTE
 
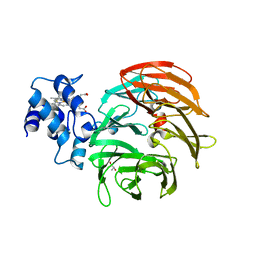 | | Dihydro-heme d1 dehydrogenase NirN in complex with DHE | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Cytochrome c, HEME C | | Authors: | Kluenemann, T, Preuss, A, Layer, G, Blankenfeldt, W. | | Deposit date: | 2019-05-23 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structure of Dihydro-Heme d1Dehydrogenase NirN from Pseudomonas aeruginosa Reveals Amino Acid Residues Essential for Catalysis.
J.Mol.Biol., 431, 2019
|
|
8PJM
 
 | |
8TOK
 
 | |
6S21
 
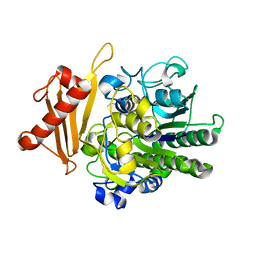 | | Metabolism of multiple glycosaminoglycans by bacteroides thetaiotaomicron is orchestrated by a versatile core genetic locus (BT33494S-sulf) | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid, CALCIUM ION, Endo-4-O-sulfatase | | Authors: | Ndeh, D, Basle, A, Strahl, H, Henrissat, B, Terrapon, N, Cartmell, A. | | Deposit date: | 2019-06-19 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Metabolism of multiple glycosaminoglycans by Bacteroides thetaiotaomicron is orchestrated by a versatile core genetic locus.
Nat Commun, 11, 2020
|
|
4CWR
 
 | | Human HSP90 alpha N-terminal domain in complex with an Aminotriazoloquinazoline inhibitor | | Descriptor: | 5-(1,3-benzodioxol-5-ylmethyl)-10-fluoro[1,2,4]triazolo[1,5-c]quinazolin-2-amine, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Casale, E, Amboldi, N, Brasca, G, Caronni, D, Colombo, N, Dalvit, C, Felder, E.R, Fogliatto, G, Isacchi, A, Mantegani, S, Polucci, P, Riceputi, L, Sola, F, Visco, C, Zuccotto, F, Casuscelli, F. | | Deposit date: | 2014-04-03 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fragment-Based Hit Discovery and Structure-Based Optimization of Aminotriazoloquinazolines as Novel Hsp90 Inhibitors.
Bioorg.Med.Chem., 22, 2014
|
|
8TPR
 
 | |
8TPQ
 
 | |
5MB4
 
 | |
8TPM
 
 | |
4C4K
 
 | | Crystal structure of the titin M10-Obscurin Ig domain 1 complex | | Descriptor: | 1,2-ETHANEDIOL, OBSCURIN, TITIN | | Authors: | Pernigo, S, Fukuzawa, A, Gautel, M, Steiner, R.A. | | Deposit date: | 2013-09-05 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of the Human Titin:Obscurin Complex Reveals a Conserved Yet Specific Muscle M-Band Zipper Module.
J.Mol.Biol., 427, 2015
|
|
6S3G
 
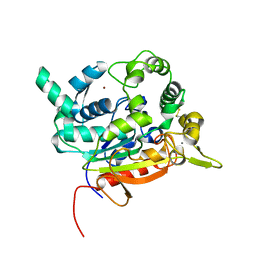 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 variant A187C/F291C | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Gihaz, S, Bash, Y, Rush, I, Shahar, A, Pazy, Y, Fishman, A. | | Deposit date: | 2019-06-25 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bridges to Stability: Engineering Disulfide Bonds Towards Enhanced Lipase Biodiesel Synthesis
Chemcatchem, 2019
|
|
8TPS
 
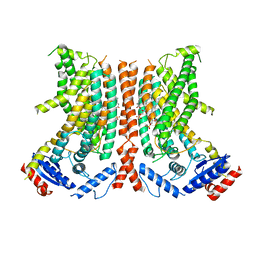 | |
8Q7C
 
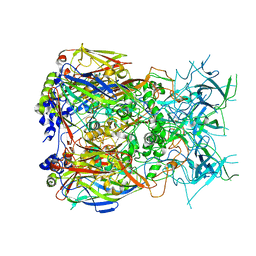 | | Cryo-EM structure of Adenovirus C5 hexon | | Descriptor: | Hexon protein | | Authors: | Zoll, S, Dhillon, A. | | Deposit date: | 2023-08-16 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into the interaction between adenovirus C5 hexon and human lactoferrin.
J.Virol., 98, 2024
|
|
8TPN
 
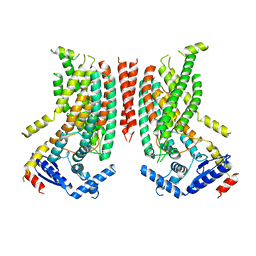 | |
5TXF
 
 | | Crystal structure of Lecithin:cholesterol acyltransferase (LCAT) in a closed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Phosphatidylcholine-sterol acyltransferase, ... | | Authors: | Manthei, K.A, Glukhova, A, Tesmer, J.J.G. | | Deposit date: | 2016-11-16 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A retractable lid in lecithin:cholesterol acyltransferase provides a structural mechanism for activation by apolipoprotein A-I.
J. Biol. Chem., 292, 2017
|
|
