6M15
 
 | | Cryo-EM structures of HKU2 spike glycoproteins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, X, Yu, J, Qiao, S, Guo, R. | | Deposit date: | 2020-02-24 | | Release date: | 2020-05-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Cryo-EM structures of HKU2 and SADS-CoV spike glycoproteins provide insights into coronavirus evolution.
Nat Commun, 11, 2020
|
|
5Y4O
 
 | | Cryo-EM structure of MscS channel, YnaI | | Descriptor: | Low conductance mechanosensitive channel YnaI | | Authors: | Zhang, Y, Yu, J. | | Deposit date: | 2017-08-04 | | Release date: | 2019-03-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A binding-block ion selective mechanism revealed by a Na/K selective channel.
Protein Cell, 9, 2018
|
|
8JL3
 
 | | membrane proteins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYL COENZYME *A, ... | | Authors: | Yu, J, Ge, J.P, Xu, R.S. | | Deposit date: | 2023-06-02 | | Release date: | 2024-05-22 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structure and mechanism of lysosome transmembrane acetylation by HGSNAT.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8JKV
 
 | | membrane proteins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Yu, J, Ge, J.P, Ruisheng, X. | | Deposit date: | 2023-06-02 | | Release date: | 2024-05-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Structure and mechanism of lysosome transmembrane acetylation by HGSNAT.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8JL1
 
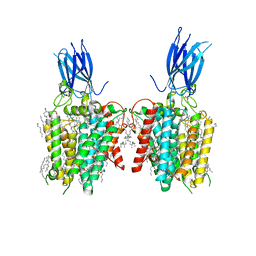 | | membrane proteins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYL COENZYME *A, ... | | Authors: | Yu, J, Ge, J.P, Xu, R.S. | | Deposit date: | 2023-06-02 | | Release date: | 2024-05-22 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure and mechanism of lysosome transmembrane acetylation by HGSNAT.
Nat.Struct.Mol.Biol., 31, 2024
|
|
3W79
 
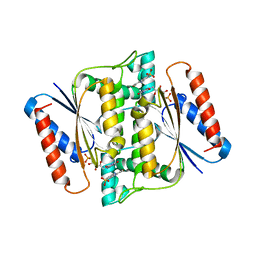 | | Crystal Structure of azoreductase AzrC in complex with sulfone-modified azo dye Orange I | | Descriptor: | 4-[(E)-(4-hydroxynaphthalen-1-yl)diazenyl]benzenesulfonic acid, FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase | | Authors: | Ogata, D, Yu, J, Ooi, T, Yao, M. | | Deposit date: | 2013-02-27 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of AzrA and of AzrC complexed with substrate or inhibitor: insight into substrate specificity and catalytic mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3UEM
 
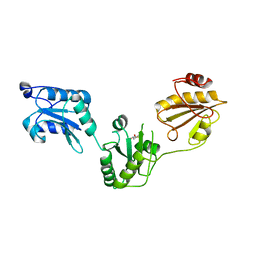 | | Crystal structure of human PDI bb'a' domains | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, Protein disulfide-isomerase | | Authors: | Yu, J, Wang, C, Huo, L, Feng, W, Wang, C.-C. | | Deposit date: | 2011-10-30 | | Release date: | 2011-11-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Human protein-disulfide isomerase is a redox-regulated chaperone activated by oxidation of domain a'
J.Biol.Chem., 287, 2012
|
|
3W77
 
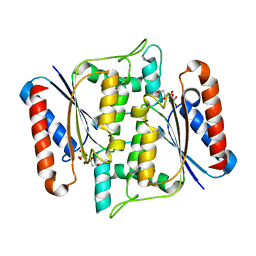 | | Crystal Structure of azoreductase AzrA | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase | | Authors: | Ogata, D, Yu, J, Ooi, T, Yao, M. | | Deposit date: | 2013-02-27 | | Release date: | 2014-02-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structures of AzrA and of AzrC complexed with substrate or inhibitor: insight into substrate specificity and catalytic mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7DCL
 
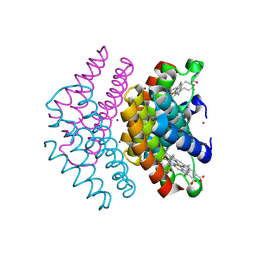 | |
7CMC
 
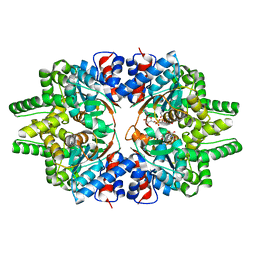 | | CRYSTAL STRUCTURE OF DEOXYHYPUSINE SYNTHASE FROM PYROCOCCUS HORIKOSHII | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Probable deoxyhypusine synthase | | Authors: | Yu, J, Gai, Z.Q, Okada, C, Yao, M. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Flexible NAD+Binding in Deoxyhypusine Synthase Reflects the Dynamic Hypusine Modification of Translation Factor IF5A.
Int J Mol Sci, 21, 2020
|
|
3D8X
 
 | | Crystal Structure of Saccharomyces cerevisiae NDPPH Dependent Thioredoxin Reductase 1 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Thioredoxin reductase 1 | | Authors: | Zhang, Z.Y, Bao, R, Yu, J, Chen, Y.X, Zhou, C.-Z. | | Deposit date: | 2008-05-26 | | Release date: | 2008-12-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Saccharomyces cerevisiae cytoplasmic thioredoxin reductase Trr1 reveals the structural basis for species-specific recognition of thioredoxin
Biochim.Biophys.Acta, 1794, 2009
|
|
6LTH
 
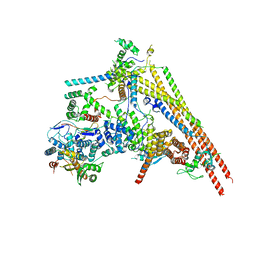 | | Structure of human BAF Base module | | Descriptor: | AT-rich interactive domain-containing protein 1A, SWI/SNF complex subunit SMARCC2, SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1, ... | | Authors: | He, S, Wu, Z, Tian, Y, Yu, Z, Yu, J, Wang, X, Li, J, Liu, B, Xu, Y. | | Deposit date: | 2020-01-22 | | Release date: | 2020-02-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of nucleosome-bound human BAF complex.
Science, 367, 2020
|
|
3K7G
 
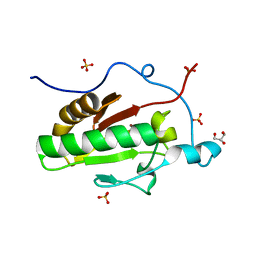 | | Crystal structure of the Indian Hedgehog N-terminal signalling domain | | Descriptor: | GLYCEROL, Indian hedgehog protein, SULFATE ION, ... | | Authors: | He, Y.-X, Kang, Y, Zhang, W.J, Yu, J, Ma, G, Zhou, C.-Z. | | Deposit date: | 2009-10-13 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the Indian Hedgehog N-terminal signalling domain
To be Published
|
|
3K7J
 
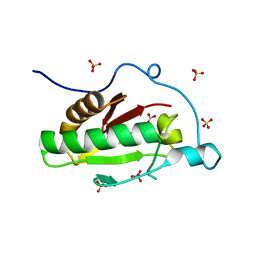 | | Crystal structure of the D100E mutant of the Indian Hedgehog N-terminal signalling domain | | Descriptor: | CARBONATE ION, Indian hedgehog protein, SULFATE ION, ... | | Authors: | He, Y.-X, Kang, Y, Zhang, W.J, Yu, J, Ma, G, Zhou, C.-Z. | | Deposit date: | 2009-10-13 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the D100E mutant of the Indian Hedgehog N-terminal signalling domain
To be Published
|
|
5JWA
 
 | | the structure of malaria PfNDH2 | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, FRAGMENT OF TRITON X-100, ... | | Authors: | Yu, Y, Yang, Y.Q, Li, X.L, Yu, J, Ge, J.P, Li, J, Rao, Y, Yang, M.J. | | Deposit date: | 2016-05-11 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.162 Å) | | Cite: | Target Elucidation by Cocrystal Structures of NADH-Ubiquinone Oxidoreductase of Plasmodium falciparum (PfNDH2) with Small Molecule To Eliminate Drug-Resistant Malaria
J. Med. Chem., 60, 2017
|
|
5X5S
 
 | |
6LTJ
 
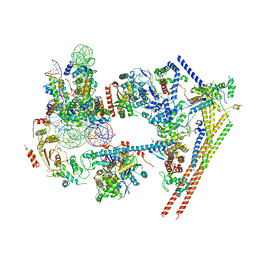 | | Structure of nucleosome-bound human BAF complex | | Descriptor: | AT-rich interactive domain-containing protein 1A, Actin, cytoplasmic 1, ... | | Authors: | He, S, Wu, Z, Tian, Y, Yu, Z, Yu, J, Wang, X, Li, J, Liu, B, Xu, Y. | | Deposit date: | 2020-01-22 | | Release date: | 2020-02-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of nucleosome-bound human BAF complex.
Science, 367, 2020
|
|
7F3A
 
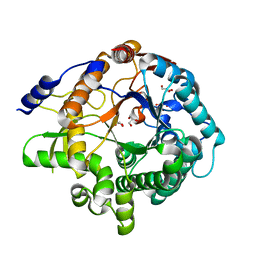 | | Arabidopsis thaliana GH1 beta-glucosidase AtBGlu42 | | Descriptor: | Beta-glucosidase 42, GLYCEROL | | Authors: | Horikoshi, S, Saburi, W, Yu, J, Yao, M. | | Deposit date: | 2021-06-16 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substrate specificity of glycoside hydrolase family 1 beta-glucosidase AtBGlu42 from Arabidopsis thaliana and its molecular mechanism.
Biosci.Biotechnol.Biochem., 86, 2022
|
|
6LIV
 
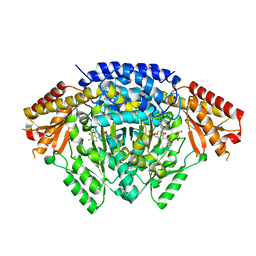 | |
4EJQ
 
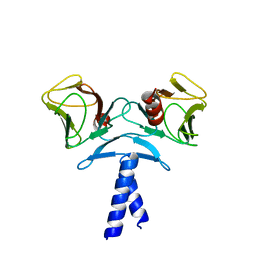 | | Crystal structure of KIF1A C-CC1-FHA | | Descriptor: | Kinesin-like protein KIF1A | | Authors: | Huo, L, Yue, Y, Ren, J, Yu, J, Liu, J, Yu, Y, Ye, F, Xu, T, Zhang, M, Feng, W. | | Deposit date: | 2012-04-06 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | The CC1-FHA Tandem as a Central Hub for Controlling the Dimerization and Activation of Kinesin-3 KIF1A
Structure, 20, 2012
|
|
6M0J
 
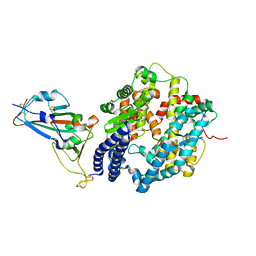 | | Crystal structure of SARS-CoV-2 spike receptor-binding domain bound with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | Authors: | Wang, X, Lan, J, Ge, J, Yu, J, Shan, S. | | Deposit date: | 2020-02-21 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor.
Nature, 581, 2020
|
|
2OWL
 
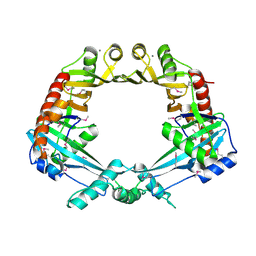 | | Crystal structure of E. coli RdgC | | Descriptor: | CALCIUM ION, Recombination-associated protein rdgC | | Authors: | Briggs, G.S, McEwan, P.A, Yu, J, Moore, T, Emsley, J, Lloyd, R.G. | | Deposit date: | 2007-02-16 | | Release date: | 2007-03-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Ring Structure of the Escherichia coli DNA-binding Protein RdgC Associated with Recombination and Replication Fork Repair.
J.Biol.Chem., 282, 2007
|
|
4DSS
 
 | | Crystal structure of peroxiredoxin Ahp1 from Saccharomyces cerevisiae in complex with thioredoxin Trx2 | | Descriptor: | Peroxiredoxin type-2, Thioredoxin-2 | | Authors: | Lian, F.M, Yu, J, Ma, X.X, Yu, X.J, Chen, Y, Zhou, C.Z. | | Deposit date: | 2012-02-19 | | Release date: | 2012-04-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Snapshots of Yeast Alkyl Hydroperoxide Reductase Ahp1 Peroxiredoxin Reveal a Novel Two-cysteine Mechanism of Electron Transfer to Eliminate Reactive Oxygen Species.
J.Biol.Chem., 287, 2012
|
|
8CU6
 
 | | Crystal structure of A2AAR-StaR2-S277-bRIL in complex with a novel A2a antagonist, LJ-4517 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R,3R,4R)-2-[(8P)-6-amino-2-(hex-1-yn-1-yl)-8-(thiophen-2-yl)-9H-purin-9-yl]oxolane-3,4-diol, Adenosine receptor A2a,Soluble cytochrome b562, ... | | Authors: | Shiriaeva, A, Park, D.-J, Kim, G, Lee, Y, Hou, X, Jarhad, D.B, Kim, G, Yu, J, Hyun, Y.E, Kim, W, Gao, Z.-G, Jacobson, K.A, Han, G.W, Stevens, R.C, Jeong, L.S, Choi, S, Cherezov, V. | | Deposit date: | 2022-05-16 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | GPCR Agonist-to-Antagonist Conversion: Enabling the Design of Nucleoside Functional Switches for the A 2A Adenosine Receptor.
J.Med.Chem., 65, 2022
|
|
8CU7
 
 | | Crystal structure of A2AAR-StaR2-bRIL in complex with a novel A2a antagonist, LJ-4517 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R,3R,4R)-2-[(8P)-6-amino-2-(hex-1-yn-1-yl)-8-(thiophen-2-yl)-9H-purin-9-yl]oxolane-3,4-diol, Adenosine receptor A2a,Soluble cytochrome b562, ... | | Authors: | Shiriaeva, A, Park, D.-J, Kim, G, Lee, Y, Hou, X, Jarhad, D.B, Kim, G, Yu, J, Hyun, Y.E, Kim, W, Gao, Z.-G, Jacobson, K.A, Han, G.W, Stevens, R.C, Jeong, L.S, Choi, S, Cherezov, V. | | Deposit date: | 2022-05-16 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | GPCR Agonist-to-Antagonist Conversion: Enabling the Design of Nucleoside Functional Switches for the A 2A Adenosine Receptor.
J.Med.Chem., 65, 2022
|
|
